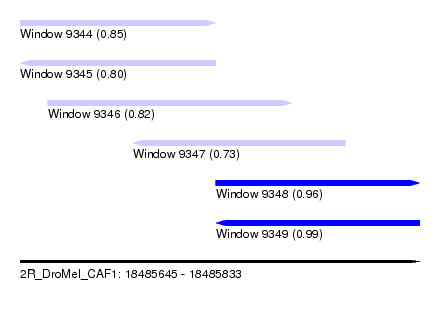
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,485,645 – 18,485,833 |
| Length | 188 |
| Max. P | 0.987243 |

| Location | 18,485,645 – 18,485,737 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 76.87 |
| Mean single sequence MFE | -21.97 |
| Consensus MFE | -16.64 |
| Energy contribution | -17.53 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18485645 92 + 20766785 GGAAAUAUAUUUAAUAGCUGUGCGAAAUUGGCGCAGCUUGUAAACACACACACACACACACAGGGCCAGAA-AGCCAAGGCAACUGCGACUGU .........((((..((((((((.......))))))))..))))...............(((((((.....-.))).((....))....)))) ( -22.90) >DroSim_CAF1 42930 82 + 1 GGAAAUAUAUUUAAUAGCUGUGCGAAAUUGGCGCAGCUUGUAAACAC----------ACACAGGGCCAGAA-AGCCAAGGCAACUGCGACUGU .........((((..((((((((.......))))))))..))))...----------..(((((((.....-.))).((....))....)))) ( -22.90) >DroAna_CAF1 40337 80 + 1 AGGAAUAUAUUUAAUAGCAGCGCGAAAUUGGCGCAGCUUGUAAACAG-------------GAGGGCCGAAGCGGGUGGGGCAGUUGCAACUGU ................(((((......(((((.(..((((....)))-------------)..)))))).((.......)).)))))...... ( -20.10) >consensus GGAAAUAUAUUUAAUAGCUGUGCGAAAUUGGCGCAGCUUGUAAACAC__________ACACAGGGCCAGAA_AGCCAAGGCAACUGCGACUGU .........((((..((((((((.......))))))))..))))...............(((((((.......)))...((....))..)))) (-16.64 = -17.53 + 0.89)

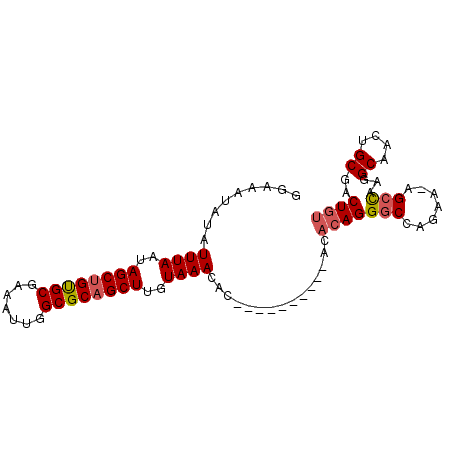
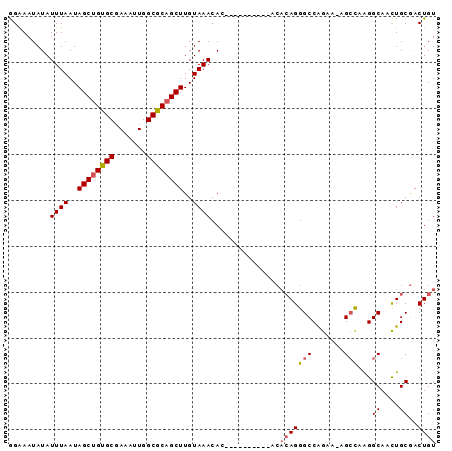
| Location | 18,485,645 – 18,485,737 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 76.87 |
| Mean single sequence MFE | -22.13 |
| Consensus MFE | -12.65 |
| Energy contribution | -14.43 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18485645 92 - 20766785 ACAGUCGCAGUUGCCUUGGCU-UUCUGGCCCUGUGUGUGUGUGUGUGUGUUUACAAGCUGCGCCAAUUUCGCACAGCUAUUAAAUAUAUUUCC (((..(((((..(((......-....))).)))))..)))....(((((((((..(((((.((.......)).)))))..))))))))).... ( -25.70) >DroSim_CAF1 42930 82 - 1 ACAGUCGCAGUUGCCUUGGCU-UUCUGGCCCUGUGU----------GUGUUUACAAGCUGCGCCAAUUUCGCACAGCUAUUAAAUAUAUUUCC ((((...(((..((....)).-..)))...))))((----------(((((((..(((((.((.......)).)))))..))))))))).... ( -23.10) >DroAna_CAF1 40337 80 - 1 ACAGUUGCAACUGCCCCACCCGCUUCGGCCCUC-------------CUGUUUACAAGCUGCGCCAAUUUCGCGCUGCUAUUAAAUAUAUUCCU .((((....))))........((....))....-------------.((((((..(((.((((.......)))).)))..))))))....... ( -17.60) >consensus ACAGUCGCAGUUGCCUUGGCU_UUCUGGCCCUGUGU__________GUGUUUACAAGCUGCGCCAAUUUCGCACAGCUAUUAAAUAUAUUUCC ((((..((....))...((((.....))))))))............(((((((..(((((.((.......)).)))))..)))))))...... (-12.65 = -14.43 + 1.78)

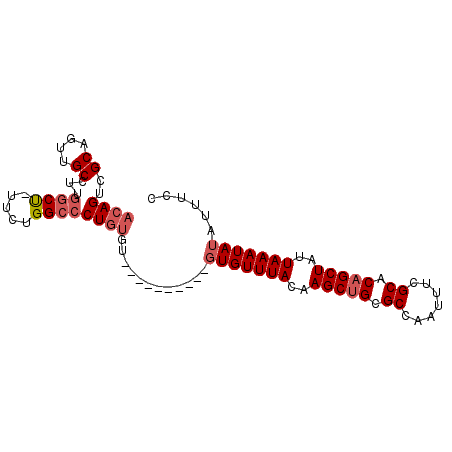
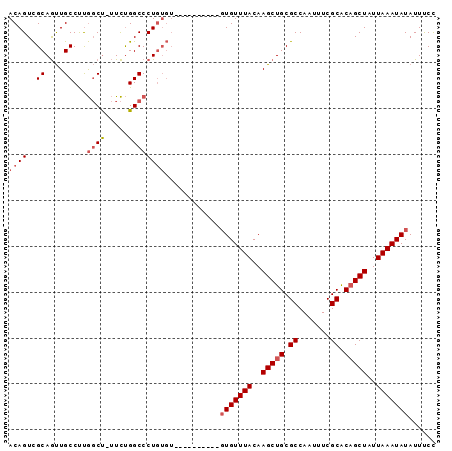
| Location | 18,485,658 – 18,485,773 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 68.11 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -15.64 |
| Energy contribution | -15.95 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18485658 115 + 20766785 AUAGCUGUGCGAAAUUGGCGCAGCUUGUAAACACACACACACACACACAGGGCCAGAA-AGCCAAGGCAACUGCGACUGUAACUGCAAUUAAAAGCCAGCCAGAUAGUGCAC-GAGU- ...((((((((...(((((...((((((......))..........(((((((.....-.))).((....))....))))............))))..)))))....)))))-.)))- ( -28.10) >DroVir_CAF1 56881 91 + 1 AUAGCAGCGCGAAAUUGGCGCAGCUUGUAAACAAG---------GCAAAGGGCGCAGC-AGC-----------------CAACAGCAAUUAAAAGCCUGCAGGGUCACGCGCUCGCUG .((((((((((....((((((((((((....))))---------((....(((.....-.))-----------------)....))..........))))...)))))))))).)))) ( -31.10) >DroSim_CAF1 42943 105 + 1 AUAGCUGUGCGAAAUUGGCGCAGCUUGUAAACAC----------ACACAGGGCCAGAA-AGCCAAGGCAACUGCGACUGUAACUGCAAUUAAAAGCCAGCCAGAUAGUGCAC-GGGU- ....(((((((...(((((...((((.(((.((.----------..(((((((.....-.))).((....))....))))...))...))).))))..)))))....)))))-))..- ( -31.00) >DroAna_CAF1 40350 103 + 1 AUAGCAGCGCGAAAUUGGCGCAGCUUGUAAACAG-------------GAGGGCCGAAGCGGGUGGGGCAGUUGCAACUGUAACAGCAAUUAAAAGCCGGCCAGACAACGCCC-CACC- .......(((....(((((.(..((((....)))-------------)..)))))).)))((((((((.((((...(((...(.((........)).)..))).))))))))-))))- ( -39.00) >consensus AUAGCAGCGCGAAAUUGGCGCAGCUUGUAAACAC__________ACACAGGGCCAAAA_AGCCAAGGCAACUGCGACUGUAACAGCAAUUAAAAGCCAGCCAGAUAACGCAC_CACU_ ......(((((...(((((...((((.(((....................(((.......)))...((....))..............))).))))..)))))....)))))...... (-15.64 = -15.95 + 0.31)

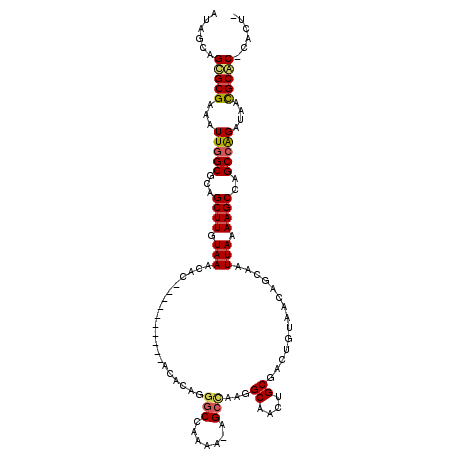
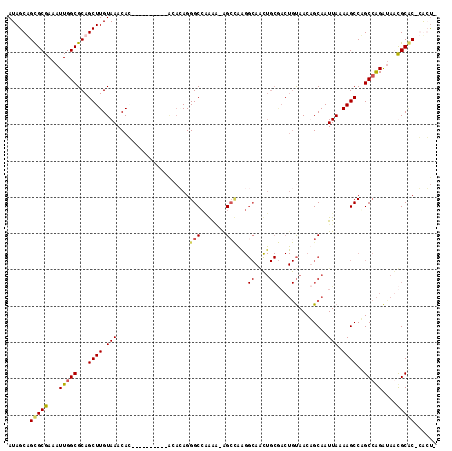
| Location | 18,485,698 – 18,485,798 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 73.15 |
| Mean single sequence MFE | -30.63 |
| Consensus MFE | -14.02 |
| Energy contribution | -15.47 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18485698 100 - 20766785 CAUACUCACACAGAUACUCACGCACACUCGUGCACUAUCUGGCUGGCUUUUAAUUGCAGUUACAGUCGCAGUUGCCUUGGCU-UUCUGGCCCUGUGUGUGU ...((.(((((((........((((....)))).......((((((((..(((((((..........)))))))....))))-....))))))))))).)) ( -30.70) >DroSim_CAF1 42977 96 - 1 CACACUCACACAGAUACUCACGCACACCCGUGCACUAUCUGGCUGGCUUUUAAUUGCAGUUACAGUCGCAGUUGCCUUGGCU-UUCUGGCCCUGUGU---- .......((((((........((((....)))).......((((((((..(((((((..........)))))))....))))-....))))))))))---- ( -26.20) >DroAna_CAF1 40384 88 - 1 CACACACACACACACACUC------GGUGGGGCGUUGUCUGGCCGGCUUUUAAUUGCUGUUACAGUUGCAACUGCCCCACCCGCUUCGGCCCUC------- ...................------((((((((((((((((..((((........))))...)))..))))).)))))))).((....))....------- ( -35.00) >consensus CACACUCACACAGAUACUCACGCACACUCGUGCACUAUCUGGCUGGCUUUUAAUUGCAGUUACAGUCGCAGUUGCCUUGGCU_UUCUGGCCCUGUGU____ ..........((((((..((((......))))...))))))...(((....((((((..........)))))))))..((((.....)))).......... (-14.02 = -15.47 + 1.45)

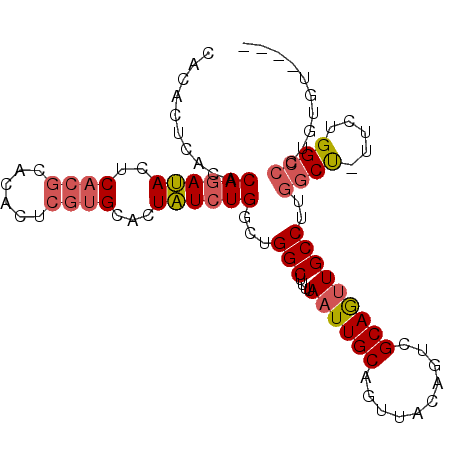
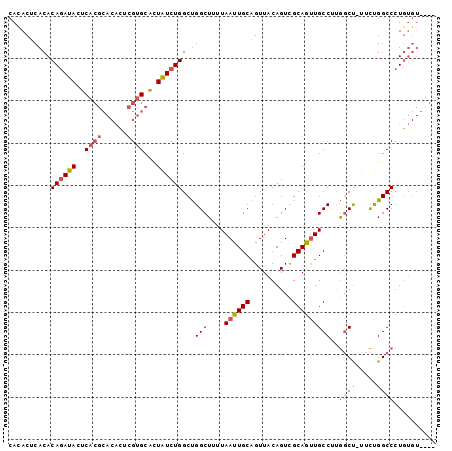
| Location | 18,485,737 – 18,485,833 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -19.98 |
| Energy contribution | -21.99 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18485737 96 + 20766785 AACUGCAAUUAAAAGCCAGCCAGAUAGUGCACGAGUGUGCGUGAGUAUCUGUGUGAGUAUGCAUUUAUGUUGUGCACACAGCUG--CAUGUGCAAAUG ...((((......(((((..((((((.(.((((......))))).))))))..)).((.(((((.......))))).)).))).--....)))).... ( -27.20) >DroSim_CAF1 43012 96 + 1 AACUGCAAUUAAAAGCCAGCCAGAUAGUGCACGGGUGUGCGUGAGUAUCUGUGUGAGUGUGCAUUUAUGCUGUGCACACAGCUG--CAUGUGCAAAUG ...((((......(((((..((((((.(.((((......))))).))))))..)).((((((((.......)))))))).))).--....)))).... ( -33.30) >DroAna_CAF1 40417 88 + 1 AACAGCAAUUAAAAGCCGGCCAGACAACGCCCCACC------GAGUGUGUGUGUGUGUGUGCAUUUAUGUUGUGCACACACUGGGCCAGAUACA---- ....((........)).((((.....(((((.(((.------..))).).))))((((((((((.......))))))))))..)))).......---- ( -29.50) >consensus AACUGCAAUUAAAAGCCAGCCAGAUAGUGCACGAGUGUGCGUGAGUAUCUGUGUGAGUGUGCAUUUAUGUUGUGCACACAGCUG__CAUGUGCAAAUG ....(((......(((..((((((((...((((......))))..)))))).))..((((((((.......)))))))).))).......)))..... (-19.98 = -21.99 + 2.00)

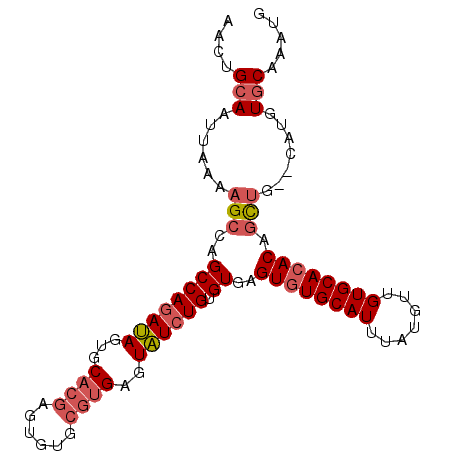
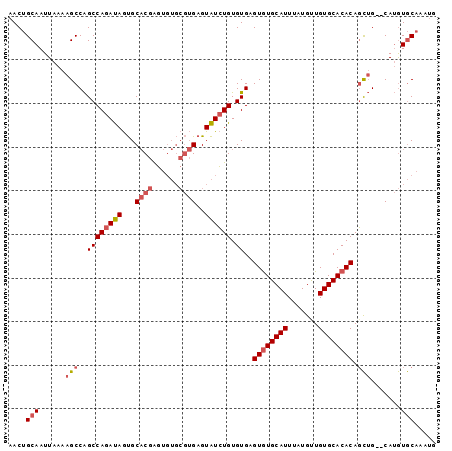
| Location | 18,485,737 – 18,485,833 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -17.10 |
| Energy contribution | -18.77 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18485737 96 - 20766785 CAUUUGCACAUG--CAGCUGUGUGCACAACAUAAAUGCAUACUCACACAGAUACUCACGCACACUCGUGCACUAUCUGGCUGGCUUUUAAUUGCAGUU ...(((((.(((--((..(((((.....)))))..)))))..(((..((((((.....((((....))))..))))))..)))........))))).. ( -25.50) >DroSim_CAF1 43012 96 - 1 CAUUUGCACAUG--CAGCUGUGUGCACAGCAUAAAUGCACACUCACACAGAUACUCACGCACACCCGUGCACUAUCUGGCUGGCUUUUAAUUGCAGUU ...(((((...(--((((.(((((((.........))))))).....((((((.....((((....))))..))))))))).)).......))))).. ( -26.80) >DroAna_CAF1 40417 88 - 1 ----UGUAUCUGGCCCAGUGUGUGCACAACAUAAAUGCACACACACACACACACUC------GGUGGGGCGUUGUCUGGCCGGCUUUUAAUUGCUGUU ----........((((.(((((((((.........))))))))).....(((....------.)))))))..........((((........)))).. ( -28.90) >consensus CAUUUGCACAUG__CAGCUGUGUGCACAACAUAAAUGCACACUCACACAGAUACUCACGCACACUCGUGCACUAUCUGGCUGGCUUUUAAUUGCAGUU ..............((((.(((((((.........))))))).....((((((..((((......))))...))))))))))((........)).... (-17.10 = -18.77 + 1.67)

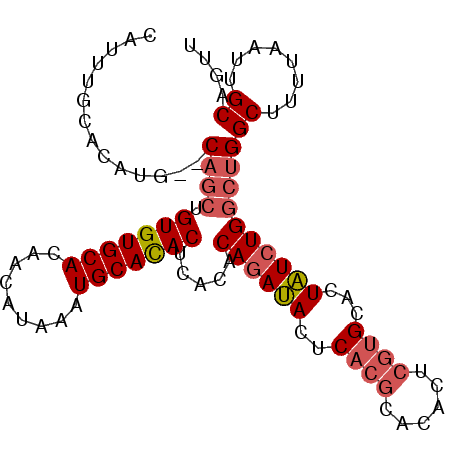
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:40 2006