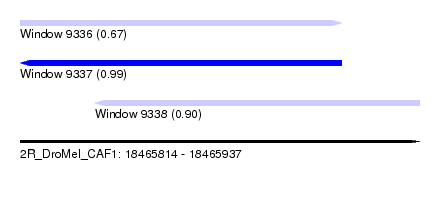
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,465,814 – 18,465,937 |
| Length | 123 |
| Max. P | 0.988469 |

| Location | 18,465,814 – 18,465,913 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -22.84 |
| Energy contribution | -23.38 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18465814 99 + 20766785 GGCUGACGCUGCAUGCAGCAGUGAAUUAUUUAUGCCAACUUGACAUUUACUUGGCCAGUAGCCUCUCUAAGCCCUGAGCUCAGUGCUCUCAGCCCAGAG (((((.((((((.....))))))..........(((((............)))))...))))).((((..((...((((.....))))...))..)))) ( -31.10) >DroSec_CAF1 27954 77 + 1 GGCUGA----------------------GUUAUGCCAACUUGACAUUUACUUGGCCAGGAGCCUCUCUAAGCCCUGAGCUCUGUGCUCUCAGCCCUGAG ((((((----------------------((((........))))...(((..(((((((.((........)))))).)))..)))...))))))..... ( -25.10) >DroSim_CAF1 28182 99 + 1 GGCUGACGCUGCAUGCAGCAGUGAAUUAUUUAUGCCAACUUGACAUUUACUUGGCCAGGAGCCUCUCUAAGCCCUGAGCUCUGUGCUCUCAGCCCUGAG ((((((((((((.....))))))........................(((..(((((((.((........)))))).)))..)))...))))))..... ( -31.90) >DroEre_CAF1 27953 99 + 1 GGCUGGCGCUGCAUGCAGCUGUGAAUUAUUUAUGCCAACUUGACAUUUACUUGGCCAGGAGCCUCUCUAAGCCCUGAGCUUUGUGCUCUCAGCUCUGAG (((((((((((....)))).((((.....)))))))..((((.((......))..)))))))).(((..(((...((((.....))))...)))..))) ( -32.20) >DroYak_CAF1 28701 99 + 1 GGCUGAGGCUGCAUGCAGCAGUGAAUUAUUUAUGCCAACUUGGCAUUUACUUGGCCAGGAGCCUCUCUAAGCCCCGAGCUUUGAGCUCUCAGUACUGAG (((..(((((((.....))))).........(((((.....)))))...))..))).(((((.((...((((.....)))).))))))).......... ( -34.10) >consensus GGCUGACGCUGCAUGCAGCAGUGAAUUAUUUAUGCCAACUUGACAUUUACUUGGCCAGGAGCCUCUCUAAGCCCUGAGCUCUGUGCUCUCAGCCCUGAG ((((..((((((.....))))))..........(((((............)))))....)))).(((...((...((((.....))))...))...))) (-22.84 = -23.38 + 0.54)

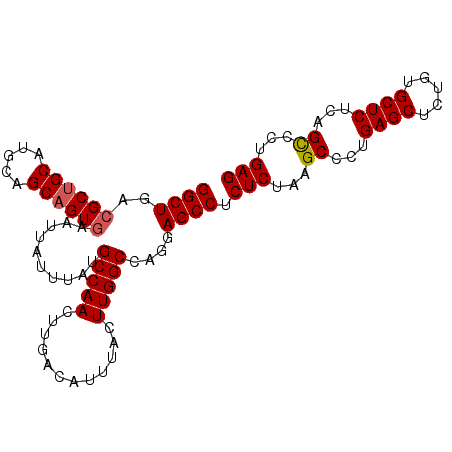
| Location | 18,465,814 – 18,465,913 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -28.36 |
| Energy contribution | -28.84 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18465814 99 - 20766785 CUCUGGGCUGAGAGCACUGAGCUCAGGGCUUAGAGAGGCUACUGGCCAAGUAAAUGUCAAGUUGGCAUAAAUAAUUCACUGCUGCAUGCAGCGUCAGCC (((((((((..((((.....))))..))))))))).((((((..(((((.(........).)))))..............((((....)))))).)))) ( -37.60) >DroSec_CAF1 27954 77 - 1 CUCAGGGCUGAGAGCACAGAGCUCAGGGCUUAGAGAGGCUCCUGGCCAAGUAAAUGUCAAGUUGGCAUAAC----------------------UCAGCC .....((((((((((.....)))((((((((....)))).))))(((((.(........).)))))....)----------------------)))))) ( -29.60) >DroSim_CAF1 28182 99 - 1 CUCAGGGCUGAGAGCACAGAGCUCAGGGCUUAGAGAGGCUCCUGGCCAAGUAAAUGUCAAGUUGGCAUAAAUAAUUCACUGCUGCAUGCAGCGUCAGCC .....((((((((((.....))))(((((((....)))).))).(((((.(........).)))))..............((((....)))).)))))) ( -33.40) >DroEre_CAF1 27953 99 - 1 CUCAGAGCUGAGAGCACAAAGCUCAGGGCUUAGAGAGGCUCCUGGCCAAGUAAAUGUCAAGUUGGCAUAAAUAAUUCACAGCUGCAUGCAGCGCCAGCC (((.(((((..((((.....))))..))))).))).((((...(((.......(((((.....)))))............((((....))))))))))) ( -34.50) >DroYak_CAF1 28701 99 - 1 CUCAGUACUGAGAGCUCAAAGCUCGGGGCUUAGAGAGGCUCCUGGCCAAGUAAAUGCCAAGUUGGCAUAAAUAAUUCACUGCUGCAUGCAGCCUCAGCC .......(((((.(((....(((.(((((((....))))))).)))..)))..(((((.....)))))............((((....))))))))).. ( -31.60) >consensus CUCAGGGCUGAGAGCACAGAGCUCAGGGCUUAGAGAGGCUCCUGGCCAAGUAAAUGUCAAGUUGGCAUAAAUAAUUCACUGCUGCAUGCAGCGUCAGCC (((.(((((..((((.....))))..))))).))).((((....(((((.(........).)))))..............((((....))))...)))) (-28.36 = -28.84 + 0.48)



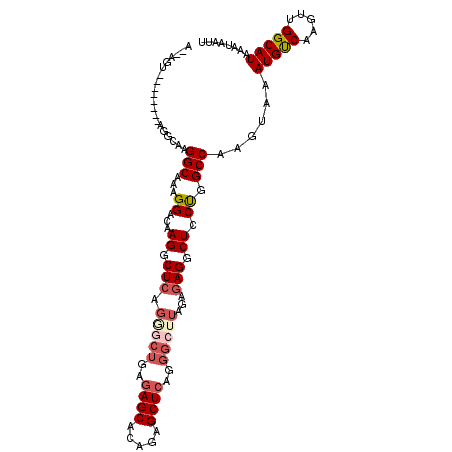
| Location | 18,465,837 – 18,465,937 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.48 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -20.75 |
| Energy contribution | -22.02 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18465837 100 - 20766785 A--AGU--------AGGCAAGGCAAAGGACAAGGCUCUGGGCUGAGAGCACUGAGCUCAGGGCUUAGAGAGGCUACUGGCCAAGUAAAUGUCAAGUUGGCAUAAAUAAUU .--...--------......(.(((..((((((((((((((((.((....)).)))))))))))).....(((.....))).......))))...))).).......... ( -35.20) >DroSec_CAF1 27960 95 - 1 A--AGU--------AGGCAAGGCAAAGGACAAGGCUCAGGGCUGAGAGCACAGAGCUCAGGGCUUAGAGAGGCUCCUGGCCAAGUAAAUGUCAAGUUGGCAUAAC----- .--...--------......(((..(((((....(((..((((..((((.....))))..))))..)))..).)))).)))......(((((.....)))))...----- ( -29.70) >DroSim_CAF1 28205 100 - 1 A--AGU--------AGGCAAGGCAAAGGACAAGGCUCAGGGCUGAGAGCACAGAGCUCAGGGCUUAGAGAGGCUCCUGGCCAAGUAAAUGUCAAGUUGGCAUAAAUAAUU .--...--------......(((..(((((....(((..((((..((((.....))))..))))..)))..).)))).)))......(((((.....)))))........ ( -29.70) >DroEre_CAF1 27976 100 - 1 A--AGU--------AGGCAAGGCAAAGGACAAGGCUCAGAGCUGAGAGCACAAAGCUCAGGGCUUAGAGAGGCUCCUGGCCAAGUAAAUGUCAAGUUGGCAUAAAUAAUU .--...--------......(((..(((((....(((.(((((..((((.....))))..))))).)))..).)))).)))......(((((.....)))))........ ( -30.10) >DroYak_CAF1 28724 108 - 1 A--AGUAGGCAAGUAGGCAAGGCAAAGGACAAGGCUCAGUACUGAGAGCUCAAAGCUCGGGGCUUAGAGAGGCUCCUGGCCAAGUAAAUGCCAAGUUGGCAUAAAUAAUU .--....(.(((.(.((((..((.........(((((........)))))....(((.(((((((....))))))).)))...))...)))).).))).).......... ( -31.50) >DroAna_CAF1 26165 81 - 1 AGUAAU--------GGGCAAGGCAAAGGACAAGGCUCACAGCU--------------CGA-------GAAGACUGCCGGCCAAGUAAAUGUCAAGCUGGCAUAAAUAAUU .((...--------.(((..((((....((..((((..(((.(--------------(..-------...)))))..))))..))...))))..))).)).......... ( -17.50) >consensus A__AGU________AGGCAAGGCAAAGGACAAGGCUCAGGGCUGAGAGCACAGAGCUCAGGGCUUAGAGAGGCUCCUGGCCAAGUAAAUGUCAAGUUGGCAUAAAUAAUU ....................(((...((...((.(((.(((((..((((.....))))..)))))...))).)).)).)))......(((((.....)))))........ (-20.75 = -22.02 + 1.26)

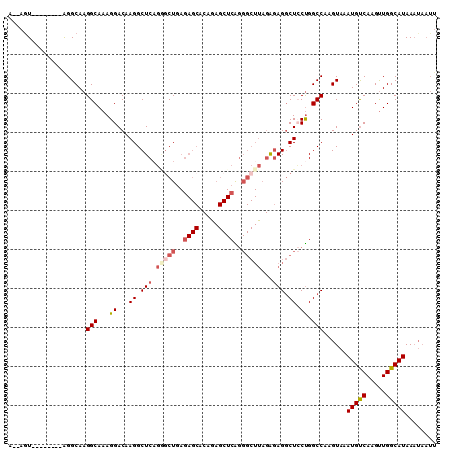
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:30 2006