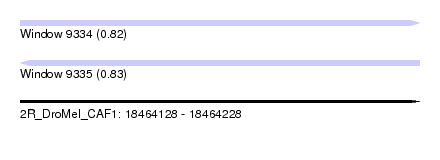
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,464,128 – 18,464,228 |
| Length | 100 |
| Max. P | 0.826755 |

| Location | 18,464,128 – 18,464,228 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -15.53 |
| Energy contribution | -17.37 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815352 |
| Prediction | RNA |
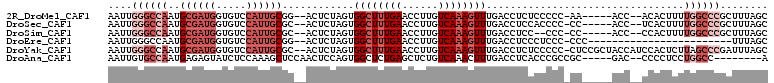
Download alignment: ClustalW | MAF
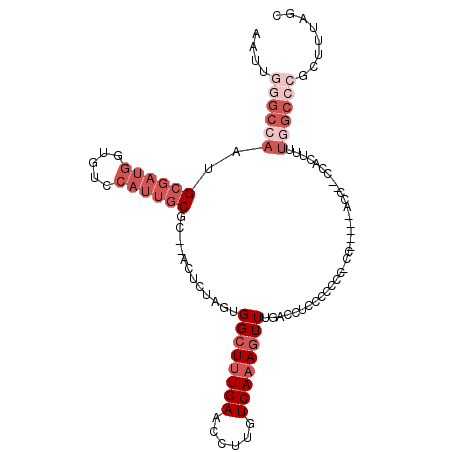
>2R_DroMel_CAF1 18464128 100 + 20766785 GCUAAAGCGGGCCAAAAGUGU--GGU-----UU-GGGGGAGAGGUCAAACUUUGACAAGGUUCAAAGCCACUAGAGU--CCGCAAUGGACACCAUCGCAUUGGCCCAAUU ((....))(((((((....((--(((-----((-..(((....((((.....))))....))).)))))))....((--((.....)))).........))))))).... ( -35.00) >DroSec_CAF1 26152 100 + 1 GCUAAAGCGGGCCAAAAGUGA--GGU-----GG-GGGGUGGAGGUCAAACUUUGACAAGGUUCAAAGCCACUAGAGU--GCGCAAUGGACACCAUCGCAUUGGCCCAAUU ((....))((((((...(((.--(((-----((-...((.((((.....)))).))...(((((..(((((....))--).))..))))).))))).))))))))).... ( -38.20) >DroSim_CAF1 26392 98 + 1 GCUAAAGCGGGCCAAAAGUGG--GGU-----GG-GGG--GGAGGUCAAACUUUGACAAGGUUCAAAGCCACUAGAGU--GCGCAAUGGACACCAUCGCAUUGGCCCAAUU ((....))((((((...(((.--(((-----((-...--....((((.....))))...(((((..(((((....))--).))..))))).))))).))))))))).... ( -38.30) >DroEre_CAF1 26214 83 + 1 GCUAAA------------------------GGG-GGGAGGGAGGUCAAACUUUGACAAGGUUCAAAGCCACUAGAGU--CCGCAAUGGACACCAUCGCAUUGGCCCAAUU ......------------------------(((-(((....((((((.....))))..(((.....))).))....)--))((.((((...)))).)).....))).... ( -20.60) >DroYak_CAF1 27035 107 + 1 GCUAAAUCGGGCUAAGAGUGGAUGGUAGCGGAG-GGGGGAGAGGUCAAACUUUGACAAGGUUCAAAGCCACUAGAGU--GCGCAAUGGACACCAUCGCAUUGGCCCAAUU ........(((((((..(((.(((((.((((((-....((....))...))))).)...(((((..(((((....))--).))..))))))))))))).))))))).... ( -34.70) >DroAna_CAF1 24059 95 + 1 U--------GGCCAGGAGGGG--GUC-----GCGGCGGGUGAGGUCAAAGUUUGACAGAGCUCAGAGCCACUGGAGUUGGAGCUUUGGAGAUACUCUCAUUGGCACAAUU (--------(((((((((((.--.((-----..(((((((...((((.....))))...))))...))).(..((((....))))..).))..))))).))))).))... ( -36.40) >consensus GCUAAAGCGGGCCAAAAGUGG__GGU_____GG_GGGGGGGAGGUCAAACUUUGACAAGGUUCAAAGCCACUAGAGU__GCGCAAUGGACACCAUCGCAUUGGCCCAAUU ........(((((((..........................((((((.....))))..(((.....))).)).........((.((((...)))).)).))))))).... (-15.53 = -17.37 + 1.84)

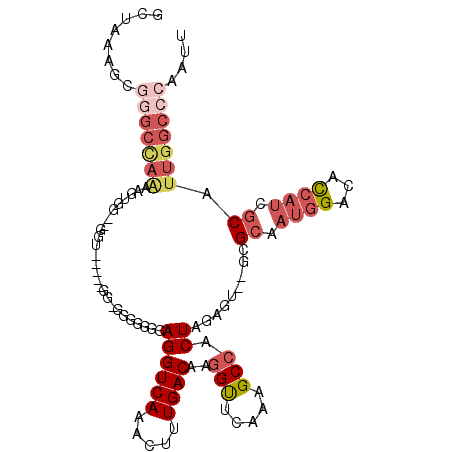
| Location | 18,464,128 – 18,464,228 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -16.05 |
| Energy contribution | -18.72 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18464128 100 - 20766785 AAUUGGGCCAAUGCGAUGGUGUCCAUUGCGG--ACUCUAGUGGCUUUGAACCUUGUCAAAGUUUGACCUCUCCCCC-AA-----ACC--ACACUUUUGGCCCGCUUUAGC ....(((((((((((((((...)))))))).--......((((.((((......((((.....))))........)-))-----)))--))....)))))))........ ( -31.94) >DroSec_CAF1 26152 100 - 1 AAUUGGGCCAAUGCGAUGGUGUCCAUUGCGC--ACUCUAGUGGCUUUGAACCUUGUCAAAGUUUGACCUCCACCCC-CC-----ACC--UCACUUUUGGCCCGCUUUAGC ....(((((((.(.((.((((..((..((.(--((....)))))..))......((((.....)))).........-.)-----)))--)).)..)))))))........ ( -29.90) >DroSim_CAF1 26392 98 - 1 AAUUGGGCCAAUGCGAUGGUGUCCAUUGCGC--ACUCUAGUGGCUUUGAACCUUGUCAAAGUUUGACCUCC--CCC-CC-----ACC--CCACUUUUGGCCCGCUUUAGC ....(((((((.(((((((...))))))).(--((....)))(((((((......))))))).........--...-..-----...--......)))))))........ ( -29.60) >DroEre_CAF1 26214 83 - 1 AAUUGGGCCAAUGCGAUGGUGUCCAUUGCGG--ACUCUAGUGGCUUUGAACCUUGUCAAAGUUUGACCUCCCUCCC-CCC------------------------UUUAGC ....(((.....(((((((...)))))))((--(.((....((((((((......)))))))).))..))).....-)))------------------------...... ( -21.40) >DroYak_CAF1 27035 107 - 1 AAUUGGGCCAAUGCGAUGGUGUCCAUUGCGC--ACUCUAGUGGCUUUGAACCUUGUCAAAGUUUGACCUCUCCCCC-CUCCGCUACCAUCCACUCUUAGCCCGAUUUAGC ((((((((.((((.(((((((......((((--((....)))(((((((......)))))))..............-...))))))))))))...)).)))))))).... ( -28.30) >DroAna_CAF1 24059 95 - 1 AAUUGUGCCAAUGAGAGUAUCUCCAAAGCUCCAACUCCAGUGGCUCUGAGCUCUGUCAAACUUUGACCUCACCCGCCGC-----GAC--CCCCUCCUGGCC--------A ...((.((((.((.((((.........))))))......(((((..((((....((((.....))))))))...)))))-----...--.......)))))--------) ( -23.10) >consensus AAUUGGGCCAAUGCGAUGGUGUCCAUUGCGC__ACUCUAGUGGCUUUGAACCUUGUCAAAGUUUGACCUCCCCCCC_CC_____ACC__CCACUUUUGGCCCGCUUUAGC ....((((((..((((((.....))))))............((((((((......)))))))).................................))))))........ (-16.05 = -18.72 + 2.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:27 2006