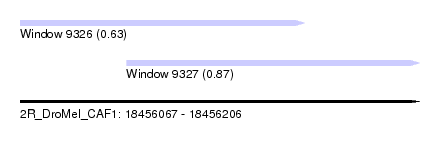
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,456,067 – 18,456,206 |
| Length | 139 |
| Max. P | 0.872675 |

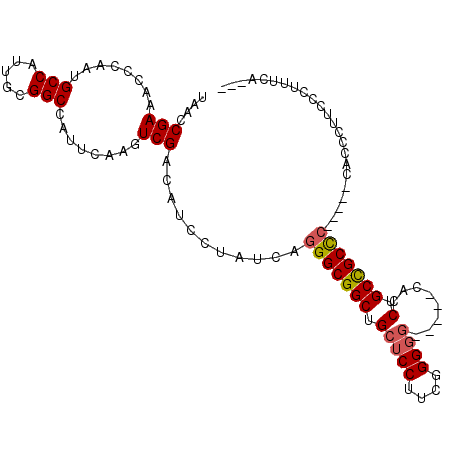
| Location | 18,456,067 – 18,456,166 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 83.76 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -20.28 |
| Energy contribution | -22.92 |
| Covariance contribution | 2.64 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18456067 99 + 20766785 UAACCGAAACCCAAUGCCAUUGCGGCCAUUCAAGUCGACAUCCUAUCAGGGCGGCUGCUCCUUCGGGGG-----CACCUGCCGCCC-----CACCCUUCCCUUUCGCCA ....(((((...........((((((.......)))).))........(((((((((((((....))))-----))...)))))))-----..........)))))... ( -34.30) >DroSec_CAF1 18458 99 + 1 UAACCGAAACCCAAUGCCAUUGCGGCCAUUCAAGUCGACAUCCUAUCAGGGCGGCUGCUCCUUCGGGGG-----CACCUGCCGCCC-----CACACUUCCCUUUCAACU ....(((........(((.....)))........)))...........(((((((((((((....))))-----))...)))))))-----.................. ( -34.19) >DroSim_CAF1 18342 99 + 1 UAACCGAAACCCAAUGCCAUUGCGGCCAUUCAAGUCGACAUCCUAUCAGGGCGGCUGCUCCUUCGGGGG-----CACCUGCCGCCC-----CACCCUUCCCUUUCAACU ....(((........(((.....)))........)))...........(((((((((((((....))))-----))...)))))))-----.................. ( -34.19) >DroEre_CAF1 18030 81 + 1 UAACCGAAACCCAAUGCCAUUGCGGCCAUUCAAGUCGACAUCCUAUCAGGGCGGCUGCUCCUUGGGGGG-----CACCUGCCGCUC----------------------- ....(((........(((.....)))........)))...........(((((((((((((....))))-----))...)))))))----------------------- ( -27.39) >DroYak_CAF1 18539 98 + 1 UAACCGAAACCCAAUGCCAUUCCGGCCAUUCAAGUCGACAUCCUAUCAGGGCGGCUGCUCCUUCGGGGCCGCUCCACCUGCUGCUC-----CACCUUCCCCAC------ ....(((........(((.....)))........))).........(((((.(((.((.((....)))).))).).))))......-----............------ ( -23.09) >DroAna_CAF1 16694 98 + 1 UAACCGAAACCCAAUGCCAUUGCGGCCAUUCAAGUCGACAUCCUAUCAGGGCGGCAGCUCCUUCGGG-G-----CACCUGCCGCCGUGGCACACCCUCCUC--CUA--- ..............((((((((((((.......)))).)).........((((((((.(((....))-)-----...))))))))))))))..........--...--- ( -33.70) >consensus UAACCGAAACCCAAUGCCAUUGCGGCCAUUCAAGUCGACAUCCUAUCAGGGCGGCUGCUCCUUCGGGGG_____CACCUGCCGCCC_____CACCCUUCCCUUUCA___ ....(((........(((.....)))........)))...........(((((((.(((((....))))........).)))))))....................... (-20.28 = -22.92 + 2.64)


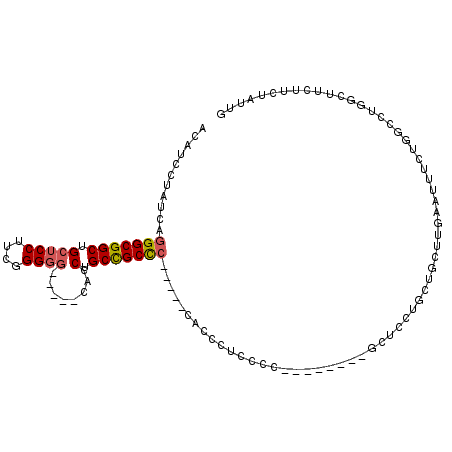
| Location | 18,456,104 – 18,456,206 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 70.36 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -13.42 |
| Energy contribution | -16.02 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18456104 102 + 20766785 ACAUCCUAUCAGGGCGGCUGCUCCUUCGGGGG-----CACCUGCCGCCC-----CACCCUUCCCUUUCGCCAGCACCUGCUGCUUGGAUUUCUGGCCUGGCUUCUUCUAUUG ...........(((((((((((((....))))-----))...)))))))-----........((....(((((..((........))....)))))..))............ ( -37.30) >DroSec_CAF1 18495 102 + 1 ACAUCCUAUCAGGGCGGCUGCUCCUUCGGGGG-----CACCUGCCGCCC-----CACACUUCCCUUUCAACUUCUCCUGCUGCUUGGAUUUCUGGCCUGGCUUCUUCUAUUG ...........(((((((((((((....))))-----))...)))))))-----...........................(((.((........)).)))........... ( -32.60) >DroEre_CAF1 18067 80 + 1 ACAUCCUAUCAGGGCGGCUGCUCCUUGGGGGG-----CACCUGCCGCUC---------------------------CUGCUGCUUGAAUUUCUGGCCUGGCUUCUACUGUUC ..........((((((((((((((....))))-----))...))))).)---------------------------))((((((.(.....).)))..)))........... ( -27.10) >DroYak_CAF1 18576 101 + 1 ACAUCCUAUCAGGGCGGCUGCUCCUUCGGGGCCGCUCCACCUGCUGCUC-----CACCUUCCCCAC------GCGUCUGUUGCUUAAAUUUCUGGCCUGGCUUCUUGUAUUG (((.....(((((.(((..........(((((.((.......)).))))-----)...........------(((.....)))........))).))))).....))).... ( -25.00) >DroAna_CAF1 16731 88 + 1 ACAUCCUAUCAGGGCGGCAGCUCCUUCGGG-G-----CACCUGCCGCCGUGGCACACCCUCCUC--CUA---GCUCC-----------CUUCU--CCCUGCUCCUUCUGCUG ........(((.((((((((.(((....))-)-----...)))))))).)))............--.((---((...-----------.....--.............)))) ( -26.30) >consensus ACAUCCUAUCAGGGCGGCUGCUCCUUCGGGGG_____CACCUGCCGCCC_____CACCCUCCCC________GCUCCUGCUGCUUGAAUUUCUGGCCUGGCUUCUUCUAUUG ...........(((((((.(((((....))))........).)))))))............................................................... (-13.42 = -16.02 + 2.60)

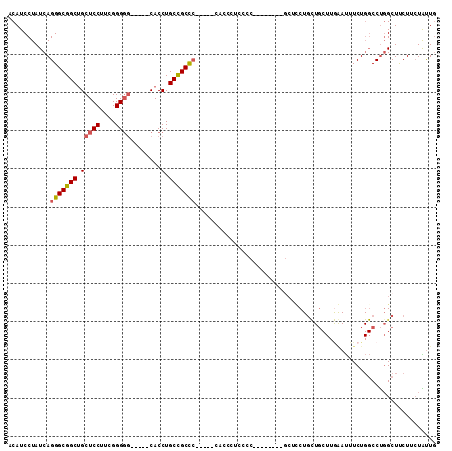
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:19 2006