
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,433,525 – 18,433,615 |
| Length | 90 |
| Max. P | 0.921333 |

| Location | 18,433,525 – 18,433,615 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -19.76 |
| Energy contribution | -19.57 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
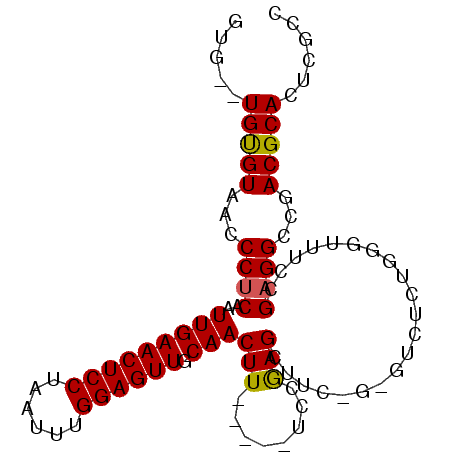
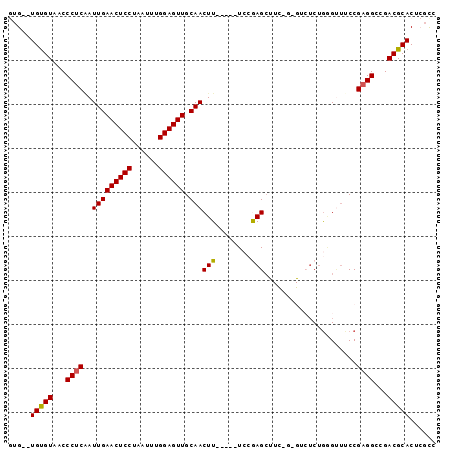
>2R_DroMel_CAF1 18433525 90 + 20766785 GUG--UGUGUAACCCUCGAUUGAACUCCUAAUUUGGAGUUGCAACUU-----UCCAAGCUUCUGCGUCUCUGUGUUCCCGAGGCCGACGCACUCGCC (.(--(((((...(((((.(((((((((......)))))).)))...-----.....((....)).............)))))...)))))).)... ( -28.90) >DroSec_CAF1 7639 81 + 1 AUG--UGUGUAACCCUCAAUUGAACUCCUAAUUUGGAGUUGCAACUU-----UCCGAGC---------UCUGGGCUUCCGAGGCUGACGCACUAGCC ..(--(((((...((((..(((((((((......)))))).)))...-----...((((---------.....))))..))))...))))))..... ( -27.50) >DroSim_CAF1 6984 81 + 1 AUG--UGUGUAACCCUCAAUUGAACUCCUAAUUUGGAGUUGCAACUU-----UCCGAGC---------UCUGGGUUUCCGAGGCUGACGCACUAGCC ..(--(((((...((((..(((((((((......)))))).)))...-----((((...---------..)))).....))))...))))))..... ( -26.00) >DroEre_CAF1 6735 90 + 1 GUG--UGUGUAACCCUCAAUUGAACUCCUAAUUUGGAGUUGCAACUU-----UCCGAGCUUCUGAGUCUCUAAGUUGCCGAGGCCGACGCACUCUCC (.(--(((((...((((.....((((((......))))))(((((((-----...(((((....)).))).))))))).))))...)))))).)... ( -34.40) >DroYak_CAF1 6796 92 + 1 GUGUGUGUGUAACCCUCAAUUGAACUCCUAAUUUGGAGUUGCAACUU-----UCCGAGAUUCAGAGUCUCUGGGUUUCCGAGGCCGACGCACUCGUG ..(.((((((...((((..(((((((((......)))))).)))...-----...(((((((((.....))))))))).))))...)))))).)... ( -34.90) >DroAna_CAF1 7243 94 + 1 GUG--UGCGUAGCCCCCCAUUGAACUCCUAAUUUGGAGUUGCAACUUGUAAUUGAAAGCCUUCCCGACGCCGAG-ACCCGAGGCCGACGCACUCGUC (.(--(((((.(((.....(((((((((......)))))).)))((((...(((..........)))...))))-......)))..)))))).)... ( -25.80) >consensus GUG__UGUGUAACCCUCAAUUGAACUCCUAAUUUGGAGUUGCAACUU_____UCCGAGCUUC_G_GUCUCUGGGUUUCCGAGGCCGACGCACUCGCC .....(((((...((((..(((((((((......)))))).)))(((........))).....................))))...)))))...... (-19.76 = -19.57 + -0.19)

| Location | 18,433,525 – 18,433,615 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -19.11 |
| Energy contribution | -18.95 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
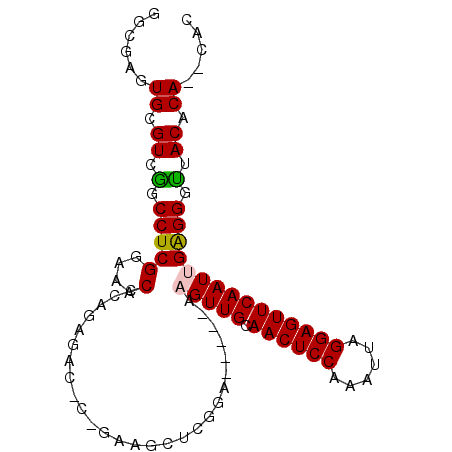
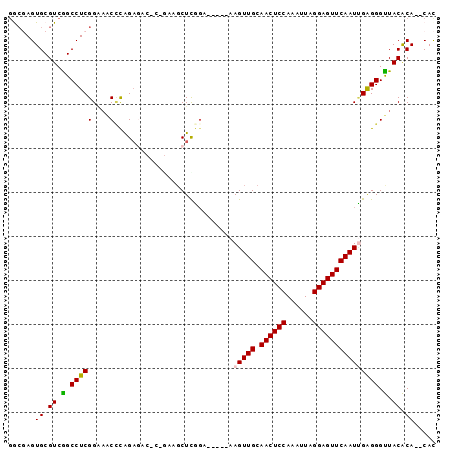
>2R_DroMel_CAF1 18433525 90 - 20766785 GGCGAGUGCGUCGGCCUCGGGAACACAGAGACGCAGAAGCUUGGA-----AAGUUGCAACUCCAAAUUAGGAGUUCAAUCGAGGGUUACACA--CAC .....(((.((.(.(((((.............((....)).....-----..((((.((((((......))))))))))))))).).)).))--).. ( -25.80) >DroSec_CAF1 7639 81 - 1 GGCUAGUGCGUCAGCCUCGGAAGCCCAGA---------GCUCGGA-----AAGUUGCAACUCCAAAUUAGGAGUUCAAUUGAGGGUUACACA--CAU .....(((.((.(.((((((....))...---------.......-----.(((((.((((((......))))))))))))))).).)).))--).. ( -25.30) >DroSim_CAF1 6984 81 - 1 GGCUAGUGCGUCAGCCUCGGAAACCCAGA---------GCUCGGA-----AAGUUGCAACUCCAAAUUAGGAGUUCAAUUGAGGGUUACACA--CAU .....(((.((.(.((((((....))...---------.......-----.(((((.((((((......))))))))))))))).).)).))--).. ( -25.30) >DroEre_CAF1 6735 90 - 1 GGAGAGUGCGUCGGCCUCGGCAACUUAGAGACUCAGAAGCUCGGA-----AAGUUGCAACUCCAAAUUAGGAGUUCAAUUGAGGGUUACACA--CAC .....(((.((.(.((((((((((((.(((.((....)))))...-----)))))))((((((......))))))....))))).).)).))--).. ( -31.90) >DroYak_CAF1 6796 92 - 1 CACGAGUGCGUCGGCCUCGGAAACCCAGAGACUCUGAAUCUCGGA-----AAGUUGCAACUCCAAAUUAGGAGUUCAAUUGAGGGUUACACACACAC ..((((.((....))))))(.(((((.((((.......))))...-----.(((((.((((((......)))))))))))..))))).)........ ( -29.30) >DroAna_CAF1 7243 94 - 1 GACGAGUGCGUCGGCCUCGGGU-CUCGGCGUCGGGAAGGCUUUCAAUUACAAGUUGCAACUCCAAAUUAGGAGUUCAAUGGGGGGCUACGCA--CAC .....((((((.((((((...(-((((....)))))................((((.((((((......))))))))))..)))))))))))--).. ( -35.90) >consensus GGCGAGUGCGUCGGCCUCGGAAACCCAGAGAC_C_GAAGCUCGGA_____AAGUUGCAACUCCAAAUUAGGAGUUCAAUUGAGGGUUACACA__CAC ......((.((.(.(((((....)...........................(((((.((((((......))))))))))))))).).)).))..... (-19.11 = -18.95 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:15 2006