| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,980,256 – 2,980,358 |
| Length | 102 |
| Max. P | 0.954873 |

| Location | 2,980,256 – 2,980,358 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.80 |
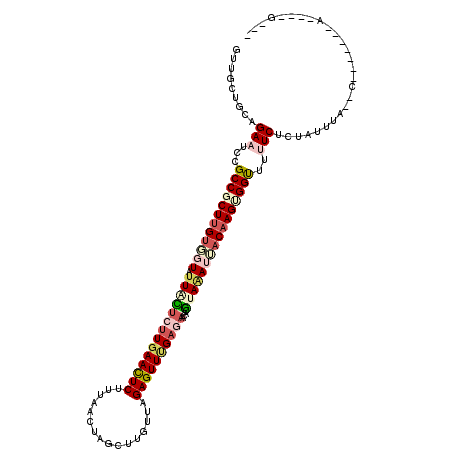
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -18.77 |
| Energy contribution | -20.47 |
| Covariance contribution | 1.70 |
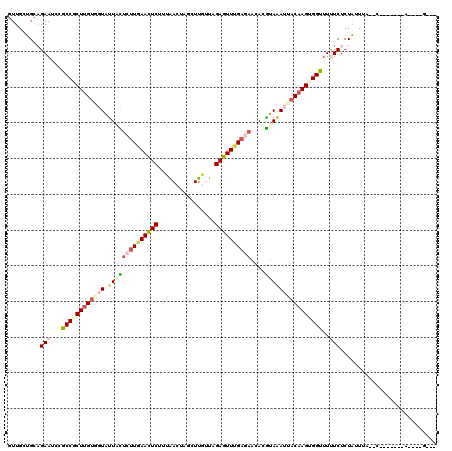
| Combinations/Pair | 1.31 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
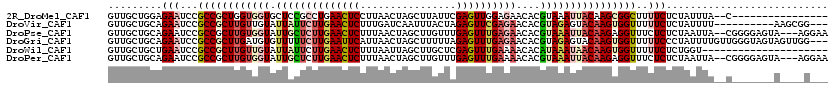
>2R_DroMel_CAF1 2980256 102 + 20766785 GUUGCUGGAGAAUCCGCCGCUGGUGGUGCUCCGCCUGAACUCCUUAACUAGCUUAUUCGAGUUGGAGAACACGUAAAUUACAAGCGGCUUUUCUCUAUUUA--C---------------- .....(((((((...(((((((((((....)))))............(((((((....))))))).................))))))..)))))))....--.---------------- ( -32.80) >DroVir_CAF1 4823 107 + 1 GUUGCUGCAGAAUCCGCCGCUUGUUGUAUUAUUCUUGAACUCUUUGAUCAAUUUACUAGAGUUCGAGAACACGUAGAGUACAAGUGGUUUUUCUCUAUUUU----------AAGCGG--- ....((((((((...((((((((((.(((..((((((((((((.(((.....)))..))))))))))))...))).)..)))))))))..)))).......----------..))))--- ( -32.00) >DroPse_CAF1 4830 115 + 1 GUUGCUGCAGAAUCCGCCGCUUGUGGUAUUGCUCUUGAACUCUUUAACUAGCUUGUUUGAGUUUGAGAACACGUAAAUUACAAGAGGUUUCUCUCUAAUUA--CGGGGAGUA---AGGAA ............((((((.((((((((.(((((((..(((((...(((......))).)))))..)))....)))))))))))).)))..((((((.....--.))))))..---.))). ( -35.50) >DroGri_CAF1 4829 117 + 1 GUUGCUGCAGAAUCCGCCGCUUGAUGUGUUUUUCUUGAAUUCAUUAACUAGCUUUUUAGAGUUUGAGAACACGUAGAGUACAAGUGGUUUUUCCCUAUUUUGUUGGGUAGUAGUUGG--- ...(((((.(((...((((((((((((((...(((..(((((................)))))..)))))))))......))))))))..)))((((......))))..)))))...--- ( -31.69) >DroWil_CAF1 5294 99 + 1 GUUGCUGCUGAAUCCGCCGCUUGUUGUAUUAUUCUUGAACUCUUUAAUUAGCUUGCUCGAGUUUGAAAACACAUAAAUAACAAGUGGUUUUUCUCUGGU--------------------- ...((((..(((...((((((((((((.((((..(..(((((................)))))..)......))))))))))))))))..)))..))))--------------------- ( -23.29) >DroPer_CAF1 6890 115 + 1 GUUGCUGCAGAAUCCGCCGCUUGUGGUAUUGCUCUUGAACUCUUUAACUAGCUUGUUUGAGUUUGAAAACACGUAAAUUACAAGAGGUUUCUCUCUAAUUA--CGGGGAGUA---AGGAA ............((((((.((((((((.((((..(..(((((...(((......))).)))))..)......)))))))))))).)))..((((((.....--.))))))..---.))). ( -30.10) >consensus GUUGCUGCAGAAUCCGCCGCUUGUGGUAUUACUCUUGAACUCUUUAACUAGCUUGUUAGAGUUUGAGAACACGUAAAUUACAAGUGGUUUUUCUCUAUUUA__C_______A____G___ .........(((...((((((((((((.((((((((((((((................))))))))))....))))))))))))))))..)))........................... (-18.77 = -20.47 + 1.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:07 2006