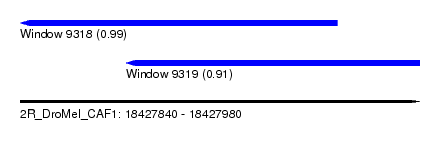
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,427,840 – 18,427,980 |
| Length | 140 |
| Max. P | 0.986582 |

| Location | 18,427,840 – 18,427,951 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -33.32 |
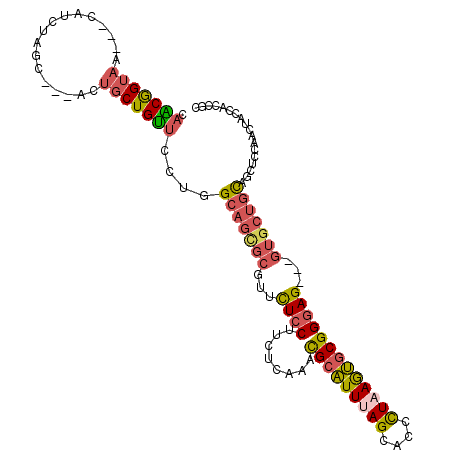
| Consensus MFE | -19.45 |
| Energy contribution | -20.31 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
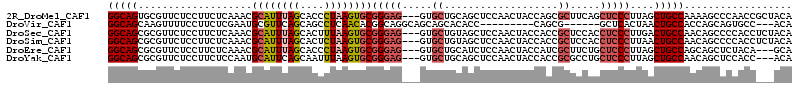
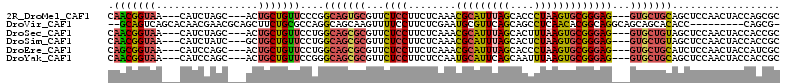
>2R_DroMel_CAF1 18427840 111 - 20766785 GGCAGUGCGUUCUCCUUCUCAAACGCAUUUAGCACCCUAAGUGCGGGAG---GUGCUGCAGCUCCAACUACCAGCGCUUCAGCUCCCUUAGCUGCCAAAAGCCCAACCGCUACA (((((((((((..........))))).....((((.....))))(((((---.((..((.(((.........)))))..)).)))))...))))))...(((......)))... ( -38.00) >DroVir_CAF1 2866 96 - 1 GGCAGCAAGUUUUCCUUCUCGAAUGCGUUCAGCAGCCUCAACACGGCAGGCAGCAGCACACC---------CAGCG------GCUCACUAACUGCCACCAGCAGUGCC---ACA ((((((..............((.(((.....)))...)).....(((((..(((.((.....---------..)).------)))......)))))....))..))))---... ( -24.50) >DroSec_CAF1 1745 111 - 1 GGCAGCGCGUUCUCCUUCUCAAACGCAUUUAGCACUUUAAGUGCGGGAG---GUGCUGUAGCUCCAACUACCACCGCUCCACCUCCCUUGACUGCCAACAGCCCCACCUCUACA (((((((((((..........))))).....((((.....))))(((((---(((....(((.............))).))))))))..).))))).................. ( -35.52) >DroSim_CAF1 1083 111 - 1 GGCAGCGCGUUCUCCUUCUCAAACGCAUUUAGCACUCUAAGUGCGGGAG---GUGCUGUAGCUCCAACUACCACCGCUCCACCUCCCUUAACUGCCAACAGCCCCACCUCUACA (((((.(((((..........))))).....((((.....))))(((((---(((....(((.............))).))))))))....))))).................. ( -35.12) >DroEre_CAF1 1098 108 - 1 GGCAGCGCGUUCUCCUUCUCAAACGCAUUUAGCACCCUAAGUGCGGGAG---GUGCUGCAUCUCCAACUACCAUCGCUUCUGCUCCCUUAGCUGCCAGCAGCUCUACA---GCA ((((((..(((............(((((((((....)))))))))((((---(((...)))))))))).......((....)).......))))))....(((....)---)). ( -36.30) >DroYak_CAF1 1104 108 - 1 GGCAGCGCGUUCUCCUUCUCCAAUGCAUUCAGCAAUUUAAGUGCGGGAG---GUGCUGCAGCUCCAACUACCACCGCGCCUGCUCCCUUAGCUGCCAACAGCUCCACC---ACA (((((((((((..........)))))....((((......((((((..(---(((.((......))..)))).)))))).))))......))))))............---... ( -30.50) >consensus GGCAGCGCGUUCUCCUUCUCAAACGCAUUUAGCACCCUAAGUGCGGGAG___GUGCUGCAGCUCCAACUACCACCGCUCCAGCUCCCUUAACUGCCAACAGCCCCACC___ACA (((((...................((((((((....))))))))(((((.....((...................)).....)))))....))))).................. (-19.45 = -20.31 + 0.86)
| Location | 18,427,877 – 18,427,980 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -33.22 |
| Consensus MFE | -21.99 |
| Energy contribution | -22.33 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
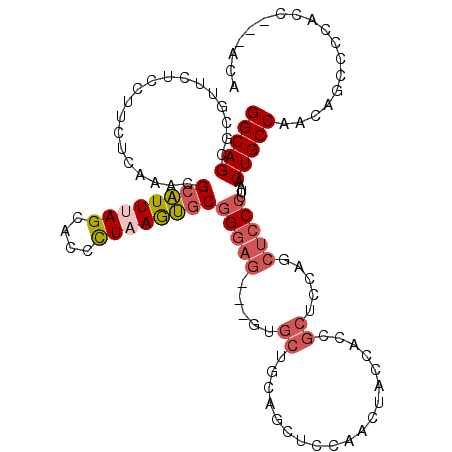
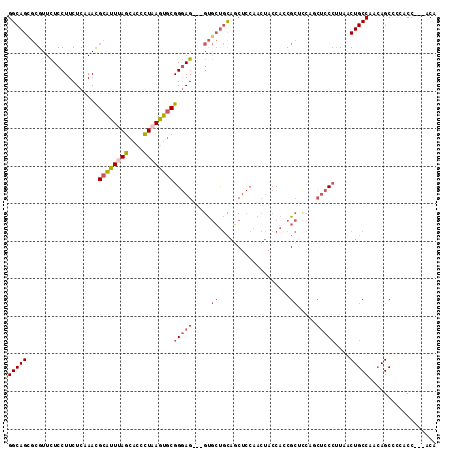
>2R_DroMel_CAF1 18427877 103 - 20766785 CAACGGUAA---CAUCUAGC---ACUGCUGUUCCCGGCAGUGCGUUCUCCUUCUCAAACGCAUUUAGCACCCUAAGUGCGGGAG---GUGCUGCAGCUCCAACUACCAGCGC ....((((.---......((---(((((((....)))))))))(((............(((((((((....)))))))))((((---.((...)).)))))))))))..... ( -38.60) >DroVir_CAF1 2895 100 - 1 --GCAGUCAGCACAACGAACGCAGCUUCUGCGCCAGGCAGCAAGUUUUCCUUCUCGAAUGCGUUCAGCAGCCUCAACACGGCAGGCAGCAGCACACC---------CAGCG- --((.(((........(((((((((((((((.....)))).)))).(((......))))))))))....(((.......))).))).)).((.....---------..)).- ( -28.40) >DroSec_CAF1 1782 103 - 1 CAACGGUAA---CAUCUAGC---ACUGCUGUUCCUGGCAGCGCGUUCUCCUUCUCAAACGCAUUUAGCACUUUAAGUGCGGGAG---GUGCUGUAGCUCCAACUACCACCGC ...((((..---.....(((---((.(((((.....)))))...........(((...(((((((((....))))))))).)))---)))))((((......)))).)))). ( -33.10) >DroSim_CAF1 1120 103 - 1 CAACGGUAA---CAUCUAUC---GCUGCUGUUCCUGGCAGCGCGUUCUCCUUCUCAAACGCAUUUAGCACUCUAAGUGCGGGAG---GUGCUGUAGCUCCAACUACCACCGC ...((((..---.......(---((((((......))))))).((...(((((.....(((((((((....)))))))))))))---).)).((((......)))).)))). ( -33.00) >DroEre_CAF1 1132 103 - 1 CAGCGGUAA---CAUCCAGC---ACUGCUGUUCCUGGCAGCGCGUUCUCCUUCUCAAACGCAUUUAGCACCCUAAGUGCGGGAG---GUGCUGCAUCUCCAACUACCAUCGC ..((((((.---..(((.((---.(((((......))))).))...............(((((((((....)))))))))))).---.)))))).................. ( -35.40) >DroYak_CAF1 1138 103 - 1 CAACGGUAA---CAUCCAGC---ACUGCUGUUCCGGGCAGCGCGUUCUCCUUCUCCAAUGCAUUCAGCAAUUUAAGUGCGGGAG---GUGCUGCAGCUCCAACUACCACCGC ...((((..---..(((.((---((((((((.....)))))(((((..........))))).............))))).)))(---(.((....)).)).......)))). ( -30.80) >consensus CAACGGUAA___CAUCUAGC___ACUGCUGUUCCUGGCAGCGCGUUCUCCUUCUCAAACGCAUUUAGCACCCUAAGUGCGGGAG___GUGCUGCAGCUCCAACUACCACCGC .(((((((.................)))))))....(((((((...((((........(((((((((....)))))))))))))...))))))).................. (-21.99 = -22.33 + 0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:12 2006