| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,396,092 – 18,396,204 |
| Length | 112 |
| Max. P | 0.548792 |

| Location | 18,396,092 – 18,396,204 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.74 |
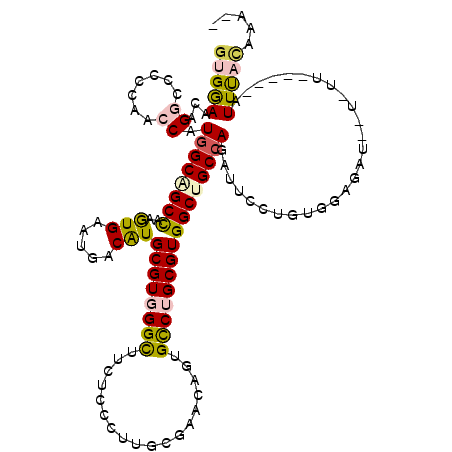
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -23.63 |
| Energy contribution | -23.89 |
| Covariance contribution | 0.26 |
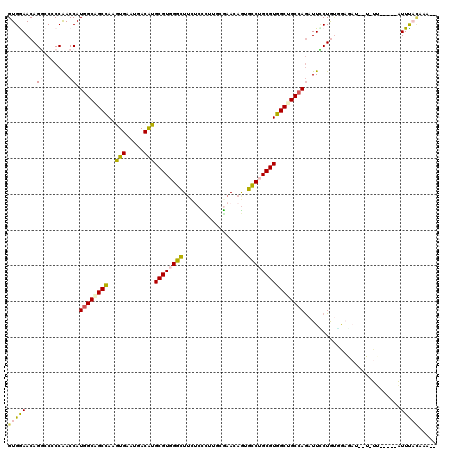
| Combinations/Pair | 1.35 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
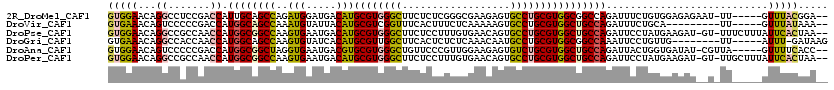
>2R_DroMel_CAF1 18396092 112 + 20766785 GUGGAACAGGCCUCCGACCAUUGCAGCCAGAUGGAUGACAUGCGUGGGCUUCUCUCGGGCGAAGAGUGCCUGCGUGGCGGCCAGAUUUCUGUGGAGAGAAU-UU-----GUUUACGGA-- (((.((((((((.((..(((((.......))))).......(((..(((..(((((....).)))).)))..))))).))))...(((((....)))))..-.)-----))))))...-- ( -39.00) >DroVir_CAF1 1843 104 + 1 GUGAAACAGUCCCCCGACCAUGGCAGCCAAAUGUAUUACAUGCGUCGGUUUCACUUUCUCAAAAAGUGCCUGCGUGGCUGCCAGAUUUCUGCA---------UU-----GUUUAUAAA-- ...(((((((..........((((((((..(((.....)))((((.((...((((((.....)))))))).)))))))))))).........)---------))-----)))).....-- ( -31.11) >DroPse_CAF1 868 116 + 1 GUGGAACAGGCCGCCAACCAUGGCGGCCAAGUGAAUGACAUGCGUGGGCUUCUCCUUUGUGAACAGUGCCUGCGUGGCUGCCAGAUUCCUAUGAAGAU-GU-UUUCUUUAUUCACUAA-- ........((((((((....)))))))).(((((((((....((((((..(((...(((....))).(((.....)))....)))..)))))).(((.-..-..))))))))))))..-- ( -41.50) >DroGri_CAF1 883 106 + 1 GUGAAACAGGCCACCAACCAUGGCAGCCAAGUGUAUCACAUGCGUUGGCUUCACUCUCUCAAACAAUGCCUGCGUGGCGGCCAAAUUCCUGUUG--------UU-----AUUU-GAUAAG ........((((.(((..((.(((((((((.(((((...)))))))))).................))))))..))).))))............--------..-----....-...... ( -26.42) >DroAna_CAF1 864 112 + 1 GUGGAACAGUCCCCCGACCAUGGCGGCUAGGUGAAUGACGUGCGUGGGCUGUUCCCGUUGGAAGAGUGUCUGCGUGGCUGCCAGAUUACUGGUGAUAU-CGUUA-----GUUUUCACC-- ..(((....)))........((((((((..((.....))..(((..((((.((((....)))).)))..)..)))))))))))((..(((..((....-))..)-----))..))...-- ( -34.50) >DroPer_CAF1 867 116 + 1 GUGGAACAGGCCGCCAACCAUGGCGGCCAAGUGAAUGACAUGCGUGGGCUUCUCCUUUGUGAACAGUGCCUGCGUGGCUGCCAGAUUCCUAUGAAGAU-GU-UUGCUUUAUUCACUAA-- ........((((((((....)))))))).(((((((((...(((..(((.......(((....))).)))..)))(((...((..(((....)))..)-).-..))))))))))))..-- ( -43.60) >consensus GUGGAACAGGCCCCCAACCAUGGCAGCCAAGUGAAUGACAUGCGUGGGCUUCUCCCUUGCGAACAGUGCCUGCGUGGCUGCCAGAUUCCUGUGGAGAU__U_UU_____AUUUACAAA__ (((((...((.......)).((((((((..(((.....)))((((((((..................))))))))))))))))...........................)))))..... (-23.63 = -23.89 + 0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:56 2006