
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,385,275 – 18,385,375 |
| Length | 100 |
| Max. P | 0.870507 |

| Location | 18,385,275 – 18,385,375 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
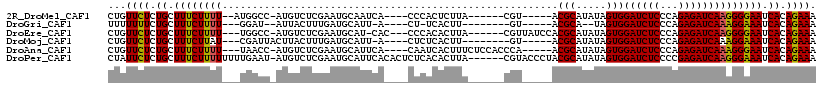
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -21.10 |
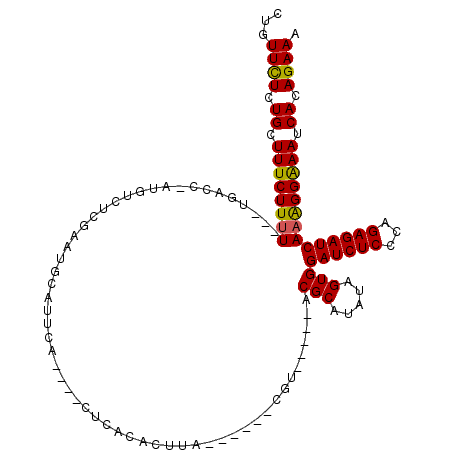
| Consensus MFE | -16.42 |
| Energy contribution | -15.95 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18385275 100 + 20766785 CUGUUCUCUGCUUUCUUUU--AUGGCC-AUGUCUCGAAUGCAAUCA----CCCACUCUUA------CGU-----ACGCAUAUAGUGGAUCUCCCAGAGAUCAAGGGGAAUCACAGAAA ((((....(((.(((...(--((....-)))....))).))).((.----(((.......------...-----.(((.....)))((((((...))))))..)))))...))))... ( -20.30) >DroGri_CAF1 29991 92 + 1 UUUUUUUCUGCUUUCUUUU---GGAU--AUUACUUUGAUGCAUU-A----CU-UCACUU--------GU-----ACGCA--UAGUGGAUCUCCCAGAGAUCAAAGGAAAUCACAGAAA ....((((((.((((((((---....--..((((...((((..(-(----(.-......--------))-----).)))--)))))((((((...))))))))))))))...)))))) ( -20.50) >DroEre_CAF1 22606 104 + 1 CUGUUCUCUGCUUUCUUUU---UGGCC-AUGUCUCGAAUGCAU-CAC---CCCACACUUA------CGUUAUCCACGCAUAUAGUGGAUCUCCCAGAGAUCAAGGGGAAUCACAGAAA ((((....(((.(((....---.(((.-..)))..))).))).-..(---(((.((((..------(((.....))).....))))((((((...))))))..))))....))))... ( -24.80) >DroMoj_CAF1 25855 97 + 1 CUGUUCUCUGCUUUCUUAU---CGAUUACUUACUUUGAUGCAUU-A----CUCUCACUU--------GU-----ACGCAUAUAGUGGAUCUCCCAGAGAUCAAAGGAAAUCACAGAAA ...((((.((.((((((..---........((((...((((..(-(----(........--------))-----).))))..))))((((((...))))))..)))))).)).)))). ( -21.80) >DroAna_CAF1 20716 105 + 1 CUGUUCUCUGCUUUCUUUU---UAACC-AUGUCUCGAAUGCAUUCA----CAAUCACUUUCUCCACCCA-----ACGCAUAUAGUGGAUCUCCCAGAGAUCAAAGGGAAUCACAGAAA ((((....(((.(((....---.....-.......))).)))....----...............(((.-----.(((.....)))((((((...))))))...)))....))))... ( -18.59) >DroPer_CAF1 29256 111 + 1 CUAUUCUCUGCUUUCUUUUUUUUGAAU-AUGUCUCGAAUGCAUUCACACUCUCACACUUA------CGUACCCUACGCAUAUAGUGGAUCUCCCCGAGAUCAAGGGAAAUCACAGAAA ...((((.((.((((((((...(((((-..((.....))..)))))........((((..------((((...)))).....))))((((((...)))))))))))))).)).)))). ( -20.60) >consensus CUGUUCUCUGCUUUCUUUU___UGACC_AUGUCUCGAAUGCAUUCA____CUCACACUUA______CGU_____ACGCAUAUAGUGGAUCUCCCAGAGAUCAAAGGAAAUCACAGAAA ...((((.((.((((((((........................................................(((.....)))((((((...)))))))))))))).)).)))). (-16.42 = -15.95 + -0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:53 2006