| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,378,749 – 18,378,844 |
| Length | 95 |
| Max. P | 0.945355 |

| Location | 18,378,749 – 18,378,844 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -12.14 |
| Energy contribution | -11.87 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
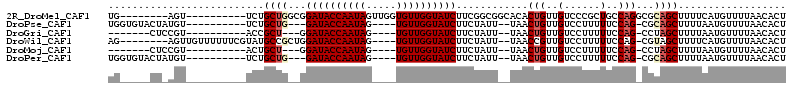
>2R_DroMel_CAF1 18378749 95 + 20766785 AGUGUUAAAACAUGAAAAGCUGCGCCUGGCAGCGGGACAACAGUGUGCCGCCGAAGAUACCAACACCAACUAUUGGUAUCCGCCAGCAGA----------ACU--------CA .((((....))))((....((((...((((.((((.(((....))).))))....((((((((.........)))))))).)))))))).----------..)--------). ( -30.90) >DroPse_CAF1 22742 93 + 1 AGUGUUAAAACAUUAAAAGCUGCG-CUGGAAAAAGGACAACAGUUA--AAUAGAAGAUACCAACA----CUAUUGGUAUC---CAGCAGA----------ACAUAGUACACCA .((((......((((....(((((-(((............))))..--.......((((((((..----...))))))))---..)))).----------...)))))))).. ( -19.50) >DroGri_CAF1 22733 86 + 1 AGUGUUAAAACAUUAAAAGCUAGG-CUGGAAAAAGGACAACAGUUA--AAUAGAAGAUACCAACA----CUAUUGGUAUCC---AGCGGU----------ACGGAG------- (((((....)))))....(((..(-(((((.....(((....))).--.........((((((..----...)))))))))---))))))----------......------- ( -17.00) >DroWil_CAF1 27460 98 + 1 AGUGUUAAAACAUGAAAAGCUACG-CUGGAAAAAGGACAACGGUUA--AAUAGAAGAUACCAACA----CUAUUGGUAUCCAGCGGCAUACGAAAAAACAACU--------CU .((((....)))).....((..((-(((((.....(((....))).--.........((((((..----...)))))))))))))))................--------.. ( -19.70) >DroMoj_CAF1 18813 86 + 1 AGUGUUAAAACAUUAAAAGCUAGG-CUGGAAAAAGGACAACAGUUA--AAUAGAAGAUACCAACA----CUAUUGGUAUCC---AGCAGU----------ACGGAG------- (((((....)))))....(((..(-(((((.....(((....))).--.........((((((..----...)))))))))---))))))----------......------- ( -17.00) >DroPer_CAF1 23354 93 + 1 AGUGUUAAAACAUUAAAAGCUGCG-CUGGAAAAAGGACAACAGUUA--AAUAGAAGAUACCAACA----CUAUUGGUAUC---CAGCAGA----------ACAUAGUACACCA .((((......((((....(((((-(((............))))..--.......((((((((..----...))))))))---..)))).----------...)))))))).. ( -19.50) >consensus AGUGUUAAAACAUUAAAAGCUACG_CUGGAAAAAGGACAACAGUUA__AAUAGAAGAUACCAACA____CUAUUGGUAUCC__CAGCAGA__________ACA_AG_____CA .((((....)))).....(((....(((............)))............((((((((.........))))))))....))).......................... (-12.14 = -11.87 + -0.28)

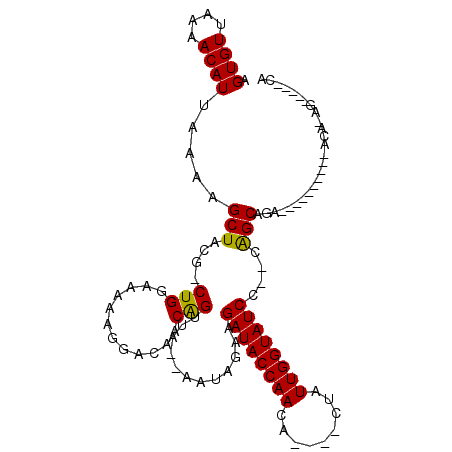
| Location | 18,378,749 – 18,378,844 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -21.67 |
| Consensus MFE | -13.50 |
| Energy contribution | -14.08 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18378749 95 - 20766785 UG--------AGU----------UCUGCUGGCGGAUACCAAUAGUUGGUGUUGGUAUCUUCGGCGGCACACUGUUGUCCCGCUGCCAGGCGCAGCUUUUCAUGUUUUAACACU .(--------(((----------(.((((((((((((((((((.....)))))))))))..(((((.(((....))).))))))))).))).)))))................ ( -35.40) >DroPse_CAF1 22742 93 - 1 UGGUGUACUAUGU----------UCUGCUG---GAUACCAAUAG----UGUUGGUAUCUUCUAUU--UAACUGUUGUCCUUUUUCCAG-CGCAGCUUUUAAUGUUUUAACACU .(((((.....((----------(.(((((---((...((((((----(..(((......)))..--..))))))).......)))))-)).)))...(((....)))))))) ( -21.50) >DroGri_CAF1 22733 86 - 1 -------CUCCGU----------ACCGCU---GGAUACCAAUAG----UGUUGGUAUCUUCUAUU--UAACUGUUGUCCUUUUUCCAG-CCUAGCUUUUAAUGUUUUAACACU -------....((----------...(((---(((...((((((----(..(((......)))..--..))))))).......)))))-)...)).................. ( -16.40) >DroWil_CAF1 27460 98 - 1 AG--------AGUUGUUUUUUCGUAUGCCGCUGGAUACCAAUAG----UGUUGGUAUCUUCUAUU--UAACCGUUGUCCUUUUUCCAG-CGUAGCUUUUCAUGUUUUAACACU ..--------(((.(((....(((.....(((((((((((((..----.))))))))).......--....(((((.........)))-)))))).....)))....)))))) ( -18.80) >DroMoj_CAF1 18813 86 - 1 -------CUCCGU----------ACUGCU---GGAUACCAAUAG----UGUUGGUAUCUUCUAUU--UAACUGUUGUCCUUUUUCCAG-CCUAGCUUUUAAUGUUUUAACACU -------....((----------...(((---(((...((((((----(..(((......)))..--..))))))).......)))))-)...)).................. ( -16.40) >DroPer_CAF1 23354 93 - 1 UGGUGUACUAUGU----------UCUGCUG---GAUACCAAUAG----UGUUGGUAUCUUCUAUU--UAACUGUUGUCCUUUUUCCAG-CGCAGCUUUUAAUGUUUUAACACU .(((((.....((----------(.(((((---((...((((((----(..(((......)))..--..))))))).......)))))-)).)))...(((....)))))))) ( -21.50) >consensus UG_____CU_AGU__________UCUGCUG__GGAUACCAAUAG____UGUUGGUAUCUUCUAUU__UAACUGUUGUCCUUUUUCCAG_CGCAGCUUUUAAUGUUUUAACACU ..........................((((...((((((((((.....))))))))))............(((..(......)..)))...)))).................. (-13.50 = -14.08 + 0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:52 2006