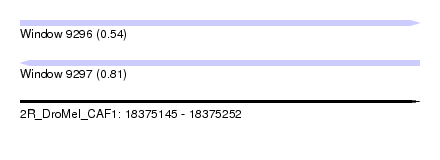
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,375,145 – 18,375,252 |
| Length | 107 |
| Max. P | 0.807161 |

| Location | 18,375,145 – 18,375,252 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 108 |
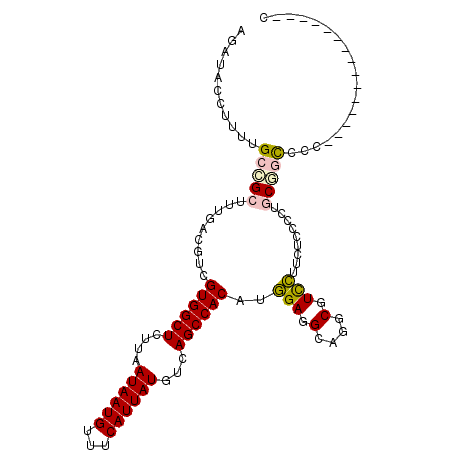
| Reading direction | forward |
| Mean pairwise identity | 81.93 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -18.23 |
| Energy contribution | -19.12 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18375145 107 + 20766785 GUGCG-AGGACGAGGGAGGGCCGCAGGGGAAAAGGACGCCUGCCUCCAUGUGGCUGACAUAAUGAAACAUUAUUAAGAGCCACGACGUCAAAGCGGCAAAAGGUAUCU .(((.-..((((..((((....(((((.(.......).))))))))).(((((((...((((((...))))))....))))))).))))...)))............. ( -35.00) >DroSec_CAF1 11956 90 + 1 G---------------GGGGCCGCAGGGGAAAAGGACGC--GCCUCCAUGUGGCUGACAUAAUGAAACAUUAUUAAGAGCCACGACGUCAAAGCGGCAA-AGGUAUCU .---------------...(((((.((((..........--..)))).(((((((...((((((...))))))....)))))))........)))))..-........ ( -26.90) >DroSim_CAF1 13119 93 + 1 G---------------GGGGCCGCAGGGGAAAAGGACGCCUGCCUCCAUGUGGCUGACAUAAUGAAACAUUAUUAAGAGCCACGACGUCAAAGCGGCAAAAGGUAUCU .---------------...(((((.((((...(((...)))..)))).(((((((...((((((...))))))....)))))))........)))))........... ( -29.90) >DroEre_CAF1 12306 99 + 1 GG------GGCGU---AGGAGGGCAGGGUAGAAGGACGCCUGCCUCCAUGUGGCUGACAUAAUGAAACAUUAUUAAGAGCCACGACGUCAAAGCGGCAAAAGGUAUCU ..------(((((---.(((((.((((((......)).))))))))).(((((((...((((((...))))))....))))))))))))................... ( -35.10) >DroWil_CAF1 21602 93 + 1 GA--------------GGGGAAGGGAUGCAGCUAGA-GGCAGCGUCUAUGUGGCUGACAUAAUGAAACAUUAUUAAGAGCCACGACGUCAAAGCUGCAAAAGGUAUCU ..--------------..........(((((((...-.(..(((((...((((((...((((((...))))))....))))))))))))..))))))).......... ( -26.00) >DroAna_CAF1 10886 102 + 1 GC---CCGGGAGG---AGGGCCGAAAGAGAGACGAAUGCCUGCCUCCAUGUGGCUGACAUAAUGAAACAUUAUUAAGAGCCACGACGUCAAAGCUGCAAAAGGUAUCU ((---(..(((((---.(((((....)..........)))).))))).(((((((...((((((...))))))....))))))).................))).... ( -29.40) >consensus G_______________AGGGCCGCAGGGGAAAAGGACGCCUGCCUCCAUGUGGCUGACAUAAUGAAACAUUAUUAAGAGCCACGACGUCAAAGCGGCAAAAGGUAUCU ...................(((((.........(((.(....).))).(((((((...((((((...))))))....)))))))........)))))........... (-18.23 = -19.12 + 0.89)

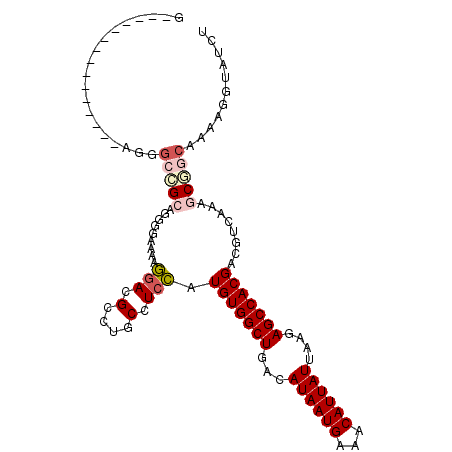
| Location | 18,375,145 – 18,375,252 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.93 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -20.00 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18375145 107 - 20766785 AGAUACCUUUUGCCGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUGGAGGCAGGCGUCCUUUUCCCCUGCGGCCCUCCCUCGUCCU-CGCAC ..............((...(((...((((((....((((((...))))))...))))))...((((.(((.((((.........).)))))).)))))))..-.)).. ( -30.70) >DroSec_CAF1 11956 90 - 1 AGAUACCU-UUGCCGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUGGAGGC--GCGUCCUUUUCCCCUGCGGCCCC---------------C ........-..(((((.....(((.((((((....((((((...))))))...)))))))))((((.--....)))).......)))))...---------------. ( -25.30) >DroSim_CAF1 13119 93 - 1 AGAUACCUUUUGCCGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUGGAGGCAGGCGUCCUUUUCCCCUGCGGCCCC---------------C ...........(((((...((((((((((((....((((((...))))))...)))))).((....))))))))..........)))))...---------------. ( -27.92) >DroEre_CAF1 12306 99 - 1 AGAUACCUUUUGCCGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUGGAGGCAGGCGUCCUUCUACCCUGCCCUCCU---ACGCC------CC ..............((.........((((((....((((((...))))))...))))))..(((((((((.((......)))))).))))).---..)).------.. ( -30.00) >DroWil_CAF1 21602 93 - 1 AGAUACCUUUUGCAGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUAGACGCUGCC-UCUAGCUGCAUCCCUUCCCC--------------UC ..........(((((((..((((((((((((....((((((...))))))...))))))...)))).....-)).)))))))..........--------------.. ( -26.10) >DroAna_CAF1 10886 102 - 1 AGAUACCUUUUGCAGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUGGAGGCAGGCAUUCGUCUCUCUUUCGGCCCU---CCUCCCGG---GC (((.((....(((.(((((..(((.((((((....((((((...))))))...))))))))))))))..)))...)).)))......((((.---......))---)) ( -27.40) >consensus AGAUACCUUUUGCCGCUUUGACGUCGUGGCUCUUAAUAAUGUUUCAUUAUGUCAGCCACAUGGAGGCAGGCGUCCUUCUCCCCUGCGGCCCC_______________C ...........(((((.........((((((....((((((...))))))...))))))..(((.(....).))).........)))))................... (-20.00 = -20.28 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:50 2006