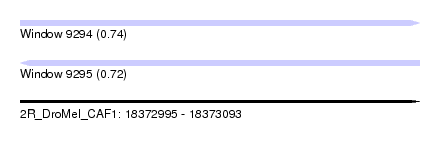
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,372,995 – 18,373,093 |
| Length | 98 |
| Max. P | 0.744160 |

| Location | 18,372,995 – 18,373,093 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.01 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
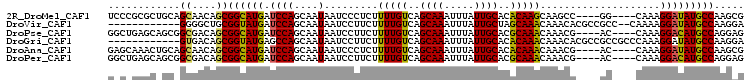
>2R_DroMel_CAF1 18372995 98 + 20766785 CGCUUGGCAUAUCCUUUG----CC----GGCUUGCUUGUGUGCAAUAAAUUUGCUGACAAAAGAGGGAUUAUUGCUGGAUCAUGCCGCUGUUGCUGCAGCGCGGGA (((..(((((((((...(----(.----..((..(((.(((((((.....))))..))).)))..))......)).)))).)))))(((((....))))))))... ( -33.10) >DroVir_CAF1 12348 92 + 1 UCCUUGGCAUAUCCUUUUG--GGCGGCGUGUUUGUUUGCUAGCAAUAAAUUUGCUGACAAAAGAAGGAUUAUUGCUGGAUCAUACCGCAGCCCC------------ ..................(--(((.(((.((.((....((((((((((.(((.((......)).))).))))))))))..)).))))).)))).------------ ( -28.60) >DroPse_CAF1 17177 98 + 1 CUCCUGGCAUGUCCUUUG----GU----CGUUUGUUUGCGUGCAAUAAAUUUGCUGACAAAAGAAGGAUUAUUGCUGGAUCAUGCCGCUGUCGCCGCUGCUCAGCC .....(((((((((..((----((----(.(((.((((((.((((.....)))))).)))).))).))))).....))).))))))((((..((....)).)))). ( -32.30) >DroGri_CAF1 14491 94 + 1 UCCUUGGCAUAUCCUUUGGGCGGCGGCGUGUUUGUUUGUGUGCAAUAAAUUUGCUGACAAAAGAAGGAUUAUUGCUGGCUCAUACCGCUGUCAC------------ .(((.((.....))...))).(((((((.((.((...((..(((((((.(((.((......)).))).)))))))..)).)).)))))))))..------------ ( -25.50) >DroAna_CAF1 9130 98 + 1 CGCUUGGCAUAUCCUUUG----GU----CGUUUGUUUGUGUGCAAUAAAUUUGCUGACAAAAGAGGGAUUAUUGCUGGAUCAUGCCGCUGUUGCUGCAGUUUGCUC .((..(((((((((..((----((----(.(((.(((((..((((.....))))..))))).))).))))).....)))).)))))(((((....)))))..)).. ( -31.70) >DroPer_CAF1 17523 98 + 1 CUCCUGGCAUGUCCUUUG----GU----CGUUUGUUUGCGUGCAAUAAAUUUGCUGACAAAAGAAGGAUUAUUGCUGGAUCAUGCCGCUGUCGCCGCUGCUCAGCC .....(((((((((..((----((----(.(((.((((((.((((.....)))))).)))).))).))))).....))).))))))((((..((....)).)))). ( -32.30) >consensus CCCUUGGCAUAUCCUUUG____GC____CGUUUGUUUGCGUGCAAUAAAUUUGCUGACAAAAGAAGGAUUAUUGCUGGAUCAUGCCGCUGUCGC_GC_GCUC_G_C ...(..(((.(((((((.............((((((.....((((.....)))).)))))).)))))))...)))..)............................ (-18.34 = -18.01 + -0.33)


| Location | 18,372,995 – 18,373,093 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -16.16 |
| Energy contribution | -15.55 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18372995 98 - 20766785 UCCCGCGCUGCAGCAACAGCGGCAUGAUCCAGCAAUAAUCCCUCUUUUGUCAGCAAAUUUAUUGCACACAAGCAAGCC----GG----CAAAGGAUAUGCCAAGCG ...(((((((......))))((((((.(((.(((((((.......((((....)))).)))))))......((.....----.)----)...)))))))))..))) ( -30.30) >DroVir_CAF1 12348 92 - 1 ------------GGGGCUGCGGUAUGAUCCAGCAAUAAUCCUUCUUUUGUCAGCAAAUUUAUUGCUAGCAAACAAACACGCCGCC--CAAAAGGAUAUGCCAAGGA ------------.((((.(((((.((..(.((((((((.......((((....)))).)))))))).)....)).)).))).)))--)....((.....))..... ( -23.00) >DroPse_CAF1 17177 98 - 1 GGCUGAGCAGCGGCGACAGCGGCAUGAUCCAGCAAUAAUCCUUCUUUUGUCAGCAAAUUUAUUGCACGCAAACAAACG----AC----CAAAGGACAUGCCAGGAG .((((.((....))..))))((((((.(((.(((((((.......((((....)))).))))))).((........))----..----....)))))))))..... ( -29.00) >DroGri_CAF1 14491 94 - 1 ------------GUGACAGCGGUAUGAGCCAGCAAUAAUCCUUCUUUUGUCAGCAAAUUUAUUGCACACAAACAAACACGCCGCCGCCCAAAGGAUAUGCCAAGGA ------------(((...((((..((.(....)............(((((..((((.....))))..)))))....))..)))))))((...((.....))..)). ( -15.90) >DroAna_CAF1 9130 98 - 1 GAGCAAACUGCAGCAACAGCGGCAUGAUCCAGCAAUAAUCCCUCUUUUGUCAGCAAAUUUAUUGCACACAAACAAACG----AC----CAAAGGAUAUGCCAAGCG ..((........((....))((((((.(((.(((((((.......((((....)))).))))))).......(....)----..----....)))))))))..)). ( -21.60) >DroPer_CAF1 17523 98 - 1 GGCUGAGCAGCGGCGACAGCGGCAUGAUCCAGCAAUAAUCCUUCUUUUGUCAGCAAAUUUAUUGCACGCAAACAAACG----AC----CAAAGGACAUGCCAGGAG .((((.((....))..))))((((((.(((.(((((((.......((((....)))).))))))).((........))----..----....)))))))))..... ( -29.00) >consensus G_C_GAGC_GC_GCGACAGCGGCAUGAUCCAGCAAUAAUCCUUCUUUUGUCAGCAAAUUUAUUGCACACAAACAAACG____AC____CAAAGGAUAUGCCAAGAG ............((....))((((((.((((....).........(((((..((((.....))))..)))))....................)))))))))..... (-16.16 = -15.55 + -0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:48 2006