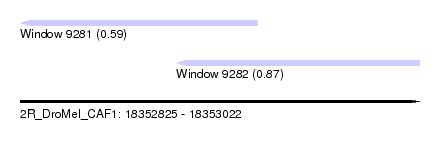
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,352,825 – 18,353,022 |
| Length | 197 |
| Max. P | 0.874656 |

| Location | 18,352,825 – 18,352,942 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -25.64 |
| Energy contribution | -26.70 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18352825 117 - 20766785 AUCAGAGGCUAAAUAAAGCGCCGGCAGUUAGCCAGGUGCUUUAUAUCAAUCGCCAGAGCCAAUUACAAUCACUUAGUUCCGGGCACUAGAUUGCUAUU---UUAUCAAAUGGCGAGGCGA ............(((((((((((((.....))).)))))))))).....(((((((.(((....((.........))....))).))...((((((((---(....)))))))))))))) ( -39.40) >DroSim_CAF1 2676 117 - 1 AUCAGAGGCUAAAUAAAGCGCCGGCAGUCGGCCAGGUACUUUAUAUCAAUCGCCAGAGCCAAUUACAAUCACUUAGUUCCGGGCACUAGACUGCUAUU---UUAUCAAAUGGCGAAGCGA .......(((..((((((..(((((.....))).))..)))))).....((((((..(((....((.........))....)))..(((....)))..---........))))))))).. ( -30.40) >DroEre_CAF1 2521 117 - 1 AUCAGAGGCUAAAUAAAGCGCCGGCACUCAGCUGGGCACUUUAUAUCAAUCGCCAGAGCCAAUUACAAUCACUUGCUUCCGGGCACUAGACUGCCUUU---UUAUCAAAUGGCAAAGCGA .......(((..((((((.((((((.....))).))).)))))).......((((((((...............))))..(((((......)))))..---........))))..))).. ( -28.96) >DroYak_CAF1 2881 120 - 1 AUCAGAGGCUAAAUAAACCGCCGGCAGUCAGCUAGACACUUUAUAUCAAUCGCCAGUGCCAAUUACAAUCACUUGGUUCCGGGCACUAGACUGCCUUUUUUUUAUCAAAUGGCAAAGCGA ....(((((..........)))(((((((.((...................)).((((((.....(((....)))......)))))).))))))).........)).............. ( -27.11) >consensus AUCAGAGGCUAAAUAAAGCGCCGGCAGUCAGCCAGGCACUUUAUAUCAAUCGCCAGAGCCAAUUACAAUCACUUAGUUCCGGGCACUAGACUGCCAUU___UUAUCAAAUGGCAAAGCGA .......(((..((((((.((((((.....))).))).)))))).....(((((((((((((..........)))))))..((((......))))..............))))))))).. (-25.64 = -26.70 + 1.06)

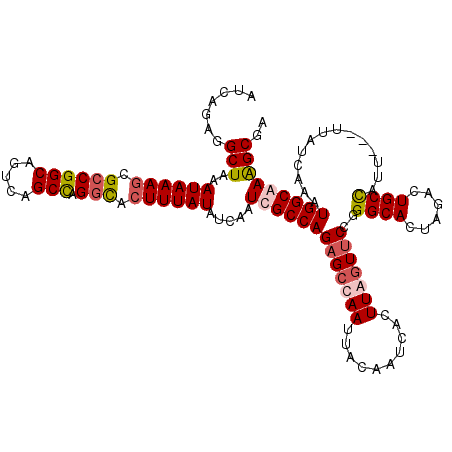
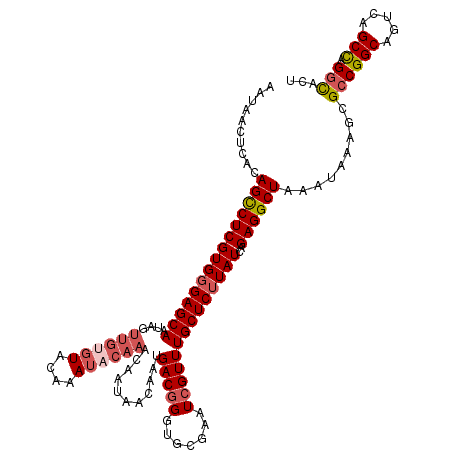
| Location | 18,352,902 – 18,353,022 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -25.76 |
| Energy contribution | -27.32 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18352902 120 - 20766785 AAUAACUCACAGUCUCGUGGGAGCAAUAGUUGUGUACAAAUACAAACAAUAACAAUGACGGGUGCGAAUCGUUUGCUCUUAUCAGAGGCUAAAUAAAGCGCCGGCAGUUAGCCAGGUGCU ..........(((((((((((((((....((((((....))))))...........(((((.......))))))))))))))..))))))......(((((((((.....))).)))))) ( -37.60) >DroSim_CAF1 2753 115 - 1 AAUAACUCACAGCCUCGUGCGAGCAAUAG-----UAUAAAUACAAACAAUAACAAUGACGGGUGCAAAUCGUUUGCUCUUAUCAGAGGCUAAAUAAAGCGCCGGCAGUCGGCCAGGUACU ..........((((((((((........)-----)))..................(((..((.((((.....)))).))..)))))))))......((..(((((.....))).))..)) ( -30.20) >DroEre_CAF1 2598 120 - 1 AAUAAUUCACAGCCUCGUGGGAGCAAUAGUUGAGUAUAAAUACAAACAAUAACAAUGACAGGUGCGAAUCGUUUGCUCUUAUCAGAGGCUAAAUAAAGCGCCGGCACUCAGCUGGGCACU ..........((((((((((((((((..(((..(((....))).))).............(((....)))..))))))))))..)))))).......((.(((((.....)))))))... ( -35.00) >DroYak_CAF1 2961 120 - 1 AAUAAUUCACAGCCUCGUGGGAGCAAUAGUUGUGUGCAAAUACAAACAAUAACAAUGACGGUUGCGAAUCGUUUGCUCUUAUCAGAGGCUAAAUAAACCGCCGGCAGUCAGCUAGACACU ..........(((((((((((((((....((((((....))))))...........((((((.....)))))))))))))))..))))))................(((.....)))... ( -31.00) >consensus AAUAACUCACAGCCUCGUGGGAGCAAUAGUUGUGUACAAAUACAAACAAUAACAAUGACGGGUGCGAAUCGUUUGCUCUUAUCAGAGGCUAAAUAAAGCGCCGGCAGUCAGCCAGGCACU ..........(((((((((((((((....((((((....))))))...........(((((.......))))))))))))))..)))))).........((((((.....))).)))... (-25.76 = -27.32 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:35 2006