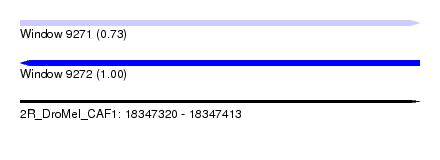
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,347,320 – 18,347,413 |
| Length | 93 |
| Max. P | 0.996334 |

| Location | 18,347,320 – 18,347,413 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.80 |
| Mean single sequence MFE | -21.51 |
| Consensus MFE | -12.61 |
| Energy contribution | -12.83 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18347320 93 + 20766785 UGUGAUAAAAAAAAUUGGCAAUGCCCACAGUAUCACUAAAUGGUA-------------------CACACCUUUAUGGUUUCUUU-GGAACAAAAAUUUAAGGCGCGACCUAUA .((((((.........(((...))).....)))))).....(((.-------------------(.(.((((.((.((((....-.))))....))..)))).).)))).... ( -16.34) >DroSim_CAF1 31341 91 + 1 UGUGAUAAAAAAA--UGGCAAUGGCCACAGUAUCACUAAAUGGUA-------------------CACACCUUUACGGUUUGUCU-UGAACAAAAAUUUAAGGCGUGACCUAUA .((((((......--((((....))))...)))))).....(((.-------------------(((.((((.....(((((..-...))))).....)))).)))))).... ( -25.60) >DroEre_CAF1 33137 110 + 1 UGUGAUAAUA---AUUGGCAAUGCCCACAGUAUCACUAAAUGAUUCGGAAAUGACAUUCAUUGCCACACCUUUAUGUUUUCUCUUGGAACAACAAUUUAAGGCGUGACCCAUA .((((((...---...(((...))).....))))))...(((..((((.((((.....)))).))((.((((..(((((((.....))).))))....)))).))))..))). ( -22.60) >consensus UGUGAUAAAAAAAAUUGGCAAUGCCCACAGUAUCACUAAAUGGUA___________________CACACCUUUAUGGUUUCUCU_GGAACAAAAAUUUAAGGCGUGACCUAUA .((((((........(((......)))...))))))............................(((.((((....(((((....)))))........)))).)))....... (-12.61 = -12.83 + 0.22)

| Location | 18,347,320 – 18,347,413 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.80 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -15.58 |
| Energy contribution | -15.25 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18347320 93 - 20766785 UAUAGGUCGCGCCUUAAAUUUUUGUUCC-AAAGAAACCAUAAAGGUGUG-------------------UACCAUUUAGUGAUACUGUGGGCAUUGCCAAUUUUUUUUAUCACA ....(((((((((((...(((((.....-..))))).....))))))))-------------------.))).....((((((.....(((...))).........)))))). ( -22.14) >DroSim_CAF1 31341 91 - 1 UAUAGGUCACGCCUUAAAUUUUUGUUCA-AGACAAACCGUAAAGGUGUG-------------------UACCAUUUAGUGAUACUGUGGCCAUUGCCA--UUUUUUUAUCACA ....(((((((((((.....((((((..-.)))))).....))))))))-------------------.))).....((((((..(((((....))))--).....)))))). ( -29.80) >DroEre_CAF1 33137 110 - 1 UAUGGGUCACGCCUUAAAUUGUUGUUCCAAGAGAAAACAUAAAGGUGUGGCAAUGAAUGUCAUUUCCGAAUCAUUUAGUGAUACUGUGGGCAUUGCCAAU---UAUUAUCACA ..(((((((((((((....((((.(((.....)))))))..))))))))))((((.....)))).))).........((((((.((..(((...)))...---)).)))))). ( -30.20) >consensus UAUAGGUCACGCCUUAAAUUUUUGUUCC_AGAGAAACCAUAAAGGUGUG___________________UACCAUUUAGUGAUACUGUGGGCAUUGCCAAUUUUUUUUAUCACA ....(((((((((((.....(((.((....)).))).....))))))))....................))).....((((((...(((......)))........)))))). (-15.58 = -15.25 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:25 2006