| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,331,247 – 18,331,346 |
| Length | 99 |
| Max. P | 0.518330 |

| Location | 18,331,247 – 18,331,346 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.45 |
| Mean single sequence MFE | -38.12 |
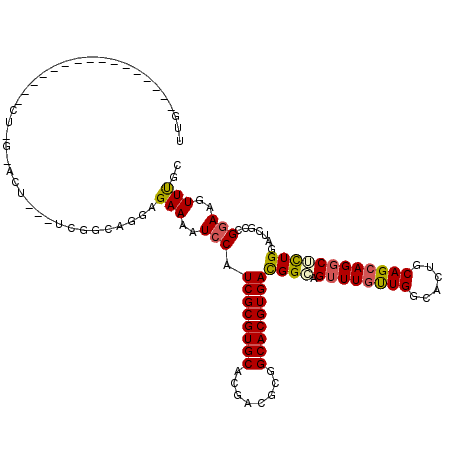
| Consensus MFE | -26.74 |
| Energy contribution | -26.72 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
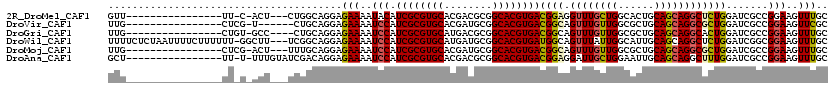
>2R_DroMel_CAF1 18331247 99 + 20766785 GUU----------------UU-C-ACU---CUGGCAGGAGAAAAUACAUCGCGUGCACGACGCGGCACGUGACGGAGGUUUGCUGGCACUGCAGCAGGCUCUGGAUCGCCGGAAGUUUGC ...----------------..-.-..(---(((((.............((((((((.((...))))))))))(((((.(((((((......))))))))))))....))))))....... ( -38.60) >DroVir_CAF1 17201 97 + 1 UUG----------------CUCG-U------CUGCAGGAGAAAAUCCAUCGCGUGCACGAUGCGGCACGUGACGGCAGUUUGUUGGCGCUGCAGCAGGCGCUGGAUCGCCGGAAGUUCGC (((----------------(...-.------..))))..(((..(((.((((((((.((...)))))))))).(((.(....(..(((((......)))))..)..)))))))..))).. ( -38.30) >DroGri_CAF1 20965 99 + 1 UUG----------------CUGU-GCC----CUGCAGGAGAAAAUCCAUCGCGUGCAUGACGCGGCACGUGACGGCAGUUUGUUGGCGCUGCAGCAGGCACUGGAUCGCCGGAAGUUUGC .((----------------(.((-(((----((((.(((.....))).((((((((........))))))))..))))......))))).)))((((((.((((....))))..)))))) ( -41.60) >DroWil_CAF1 18902 116 + 1 UUUUCUCUAAUUUUCUUUUUU-GGCUU---UCGGCAGGAGAAAAUCCAUCGCGUGCAUGAUGCGGCACGUGAUGGCAGUUUAUUGGCAUUGCAGCAGGCUCUGGAUCGGCGGAAGUUUGC ((((((((......(......-)((..---...)).))))))))..((((((((((........))))))))))(((((........))))).((((((((((......))).))))))) ( -37.20) >DroMoj_CAF1 22712 100 + 1 UUG----------------CUCG-ACU---UUUGCAGGAGAAAAUCCAUCGCGUGCACGAUGCGGCACGUGACGGCAGUUUGUUGGCGCUGCAGCAGGCGCUGGAUCGCCGGAAGUUUGC ..(----------------(..(-(((---(((((.(((.....))).((((((((.((...))))))))))..)))......(((((.(.((((....)))).).))))))))))).)) ( -37.60) >DroAna_CAF1 14342 102 + 1 GCU----------------UU-U-UUUGUAUCGACAGGAGAAAAUCCAUCGCGUGCACGACGCGGCACGUGACGGAGGAUUGCUGGAAUUGCAGCAGGCUUUGGAUCGCCGGAAGUUUGC ...----------------..-.-..((((....(((.((....(((.((((((((.((...)))))))))).)))...)).)))....))))(((((((((.(.....).))))))))) ( -35.40) >consensus UUG________________CU_G_ACU___UCGGCAGGAGAAAAUCCAUCGCGUGCACGACGCGGCACGUGACGGCAGUUUGUUGGCACUGCAGCAGGCUCUGGAUCGCCGGAAGUUUGC .......................................(((..(((.((((((((........))))))))((((.((((((((......)))))))))))).......)))..))).. (-26.74 = -26.72 + -0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:20 2006