
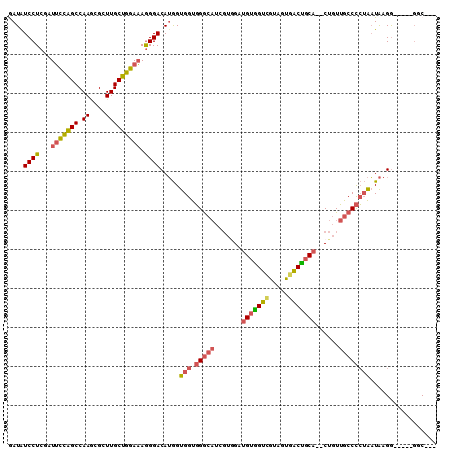
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,312,808 – 18,312,908 |
| Length | 100 |
| Max. P | 0.687949 |

| Location | 18,312,808 – 18,312,908 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -38.43 |
| Consensus MFE | -25.35 |
| Energy contribution | -25.69 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18312808 100 + 20766785 AAUGUCCUCGAUUCCAGCCAAGCGCUUGCUGGAAAGGGACAUGGUGGUGGGCAUCGUAGAUGUGGUCGUAGUGACUGCA--CUGUUGCCCCUAAUAAGG-----GGC--- .(((((((...(((((((.((....))))))))).)))))))((((.....)))).(((.((..(((.....)))..))--)))..((((((....)))-----)))--- ( -39.80) >DroSec_CAF1 6078 100 + 1 GAUAUCCUCGAUUCCAGCCAAGCGCUUGCUGGAAAGGGACAUGGUGGUGGGCAUCGUGGAUGUGGUCGUAGUGACCGCA--CUGUUGCCCCUAAUAAGG-----GGC--- ...((((.((((.((.((((.....((.((....)).)).....)))).)).)))).))))((((((.....)))))).--.....((((((....)))-----)))--- ( -40.60) >DroSim_CAF1 5459 100 + 1 GAUAUCCUCGAUUCCAGCCAAGCGCUUGCUGGAAAGGGACAUGGUGGUGGGCAUCGUGGAUGUGGUCGUAGUGACCGCA--CUGUUGCCCCUAAUAAGG-----GGC--- ...((((.((((.((.((((.....((.((....)).)).....)))).)).)))).))))((((((.....)))))).--.....((((((....)))-----)))--- ( -40.60) >DroEre_CAF1 6084 100 + 1 GAUGUCCUCGAUUCCAGCCAAGCGCUUGCUGGAAAGGGACAUGGUGGUGGGCAUGGUGGAUGUGGUUGUAGUGACUGCA--CUGUUGCCCCUAAUAAGG-----GGC--- .(((((((...(((((((.((....))))))))).)))))))..(((.(((((..(..(.((..(((.....)))..))--)..)))))))))......-----...--- ( -36.70) >DroWil_CAF1 33614 103 + 1 AAUGUCCUCAAUGUUGGCCAAACGCUUGCUGGACAAGGACAGAGCGGCUGA-------GAUGGAGUUGCAGCUGCUGCUGCCUGUGGCCUGAGCUGAGGAUAGCUGCGGC ..((((((((.....(((((...(((....)).).(((.(((((((((((.-------.((...))..)))))))).)))))).))))).....))))))))........ ( -40.10) >DroYak_CAF1 5988 100 + 1 GAUAUCCUCGAUUCCAGCCAAUCGCUUGCUGGAAAGGGACAUGGUGGUGGGCAUGGUGGAUGUGGUUGUAGUGACUGCA--CUGUUGCCCCUAAUAAGA-----GGC--- .....((((..(((((((.........)))))))..........(((.(((((..(..(.((..(((.....)))..))--)..)))))))))....))-----)).--- ( -32.80) >consensus GAUAUCCUCGAUUCCAGCCAAGCGCUUGCUGGAAAGGGACAUGGUGGUGGGCAUCGUGGAUGUGGUCGUAGUGACUGCA__CUGUUGCCCCUAAUAAGG_____GGC___ ....((((...(((((((.((....))))))))).)))).....(((.(((((.......(((((((.....)))))))......))))))))................. (-25.35 = -25.69 + 0.34)


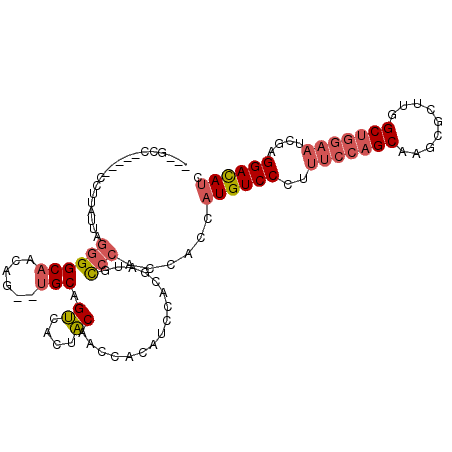
| Location | 18,312,808 – 18,312,908 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -17.42 |
| Energy contribution | -18.25 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18312808 100 - 20766785 ---GCC-----CCUUAUUAGGGGCAACAG--UGCAGUCACUACGACCACAUCUACGAUGCCCACCACCAUGUCCCUUUCCAGCAAGCGCUUGGCUGGAAUCGAGGACAUU ---(((-----(((....))))))....(--((..(((.....))))))...................(((((((.(((((((.........)))))))..).)))))). ( -33.40) >DroSec_CAF1 6078 100 - 1 ---GCC-----CCUUAUUAGGGGCAACAG--UGCGGUCACUACGACCACAUCCACGAUGCCCACCACCAUGUCCCUUUCCAGCAAGCGCUUGGCUGGAAUCGAGGAUAUC ---(((-----(((....))))))....(--((.((((.....)))).((((...))))..)))....(((((((.(((((((.........)))))))..).)))))). ( -35.60) >DroSim_CAF1 5459 100 - 1 ---GCC-----CCUUAUUAGGGGCAACAG--UGCGGUCACUACGACCACAUCCACGAUGCCCACCACCAUGUCCCUUUCCAGCAAGCGCUUGGCUGGAAUCGAGGAUAUC ---(((-----(((....))))))....(--((.((((.....)))).((((...))))..)))....(((((((.(((((((.........)))))))..).)))))). ( -35.60) >DroEre_CAF1 6084 100 - 1 ---GCC-----CCUUAUUAGGGGCAACAG--UGCAGUCACUACAACCACAUCCACCAUGCCCACCACCAUGUCCCUUUCCAGCAAGCGCUUGGCUGGAAUCGAGGACAUC ---(((-----(((....))))))...((--((....))))...........................(((((((.(((((((.........)))))))..).)))))). ( -31.30) >DroWil_CAF1 33614 103 - 1 GCCGCAGCUAUCCUCAGCUCAGGCCACAGGCAGCAGCAGCUGCAACUCCAUC-------UCAGCCGCUCUGUCCUUGUCCAGCAAGCGUUUGGCCAACAUUGAGGACAUU ..........(((((((....(((((..(((((.(((.((((..........-------.)))).))))))))((((.....))))....)))))....))))))).... ( -34.70) >DroYak_CAF1 5988 100 - 1 ---GCC-----UCUUAUUAGGGGCAACAG--UGCAGUCACUACAACCACAUCCACCAUGCCCACCACCAUGUCCCUUUCCAGCAAGCGAUUGGCUGGAAUCGAGGAUAUC ---(((-----(((....))))))...((--((....))))........((((..((((........))))((...(((((((.........)))))))..))))))... ( -26.80) >consensus ___GCC_____CCUUAUUAGGGGCAACAG__UGCAGUCACUACAACCACAUCCACGAUGCCCACCACCAUGUCCCUUUCCAGCAAGCGCUUGGCUGGAAUCGAGGACAUC ...................((((((......))).((....))................)))......((((((..(((((((.........)))))))....)))))). (-17.42 = -18.25 + 0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:14 2006