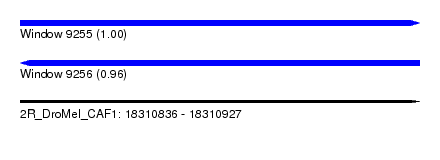
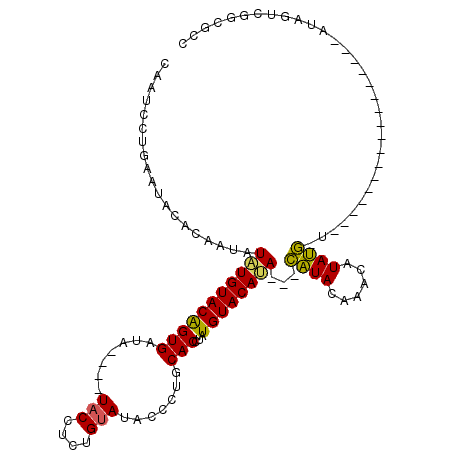
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,310,836 – 18,310,927 |
| Length | 91 |
| Max. P | 0.999280 |

| Location | 18,310,836 – 18,310,927 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 68.11 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -14.53 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.50 |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.999280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
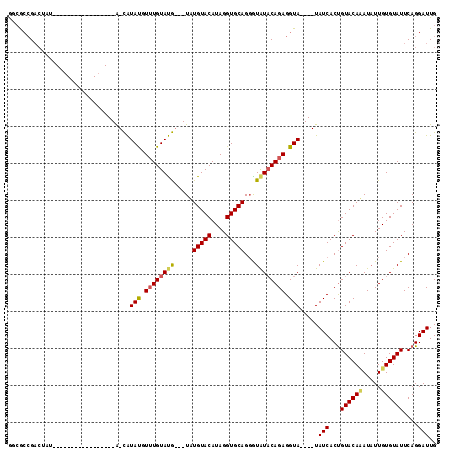
>2R_DroMel_CAF1 18310836 91 + 20766785 GGCGCCGACUAU-----------------A-CAUAUGUUUGUAUG---UAUGUACAUAGGUGCAGGGUAUACAGAGGUA----UAUCACUGUACAUAUAUUGUGUAUUCAGGAUUG ....((((.(((-----------------(-(((((((.(((((.---...)))))...(((((((((((((....)))----)))).))))))))))).)))))).)).)).... ( -27.40) >DroSec_CAF1 4108 91 + 1 GUCGCCGCCUAU-----------------A-CAUACGUUUGUAUG---UAUGUACAUAGGUGCAGCAUGUACAGAGGUA----UAUCACUGUACAAAUAUUGUGUAUUCAGGAUUG ((.((.((((((-----------------(-(((((((....)))---))))))..))))))).)).(((((((.((..----..)).)))))))...(((.((....)).))).. ( -26.70) >DroYak_CAF1 4106 113 + 1 AGGAGAGA---GAGUCUAUAUAUGUACAUGUUGUAUGUAUGUACAUAGUGUGUACAUAGGUGCAGGGUAUUCAGAGGUACAUAUAUCACCGUACACAUCUUGUGUAUUCAGGAUUG ........---.(((((((((((((((((.((((((.(((((((((...))))))))).)))))).))........))))))))))....((((((.....))))))...))))). ( -33.90) >consensus GGCGCCGACUAU_________________A_CAUAUGUUUGUAUG___UAUGUACAUAGGUGCAGGGUAUACAGAGGUA____UAUCACUGUACAAAUAUUGUGUAUUCAGGAUUG .................................(((.((((((((.....(((((....)))))...)))))))).))).....(((...((((((.....))))))....))).. (-14.53 = -15.20 + 0.67)

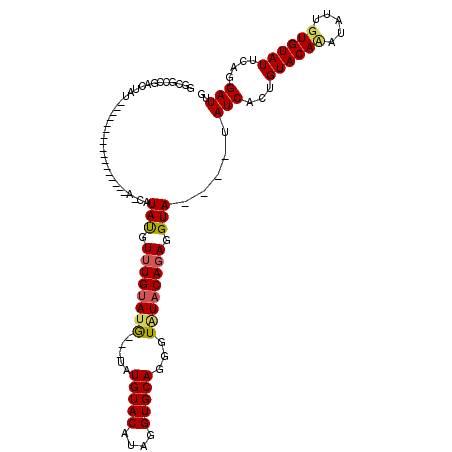
| Location | 18,310,836 – 18,310,927 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 68.11 |
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -8.90 |
| Energy contribution | -8.24 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18310836 91 - 20766785 CAAUCCUGAAUACACAAUAUAUGUACAGUGAUA----UACCUCUGUAUACCCUGCACCUAUGUACAUA---CAUACAAACAUAUG-U-----------------AUAGUCGGCGCC .....................(((((((.(...----...).)))))))....((.((..((((((((---..........))))-)-----------------)))...)).)). ( -16.80) >DroSec_CAF1 4108 91 - 1 CAAUCCUGAAUACACAAUAUUUGUACAGUGAUA----UACCUCUGUACAUGCUGCACCUAUGUACAUA---CAUACAAACGUAUG-U-----------------AUAGGCGGCGAC .....................(((((((.(...----...).)))))))((((((.....((((((((---(........)))))-)-----------------))).)))))).. ( -27.00) >DroYak_CAF1 4106 113 - 1 CAAUCCUGAAUACACAAGAUGUGUACGGUGAUAUAUGUACCUCUGAAUACCCUGCACCUAUGUACACACUAUGUACAUACAUACAACAUGUACAUAUAUAGACUC---UCUCUCCU ..(((.((......)).)))((((((((((......(((........)))....))))...)))))).(((((((..(((((.....)))))..)))))))....---........ ( -21.30) >consensus CAAUCCUGAAUACACAAUAUAUGUACAGUGAUA____UACCUCUGUAUACCCUGCACCUAUGUACAUA___CAUACAAACAUAUG_U_________________AUAGUCGGCGCC ...................(((((((((((.......(((....))).......)))...))))))))...((((......))))............................... ( -8.90 = -8.24 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:10 2006