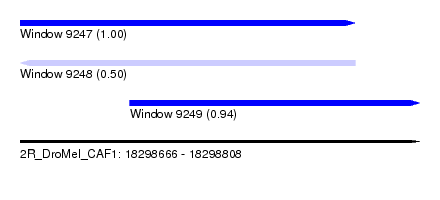
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,298,666 – 18,298,808 |
| Length | 142 |
| Max. P | 0.998772 |

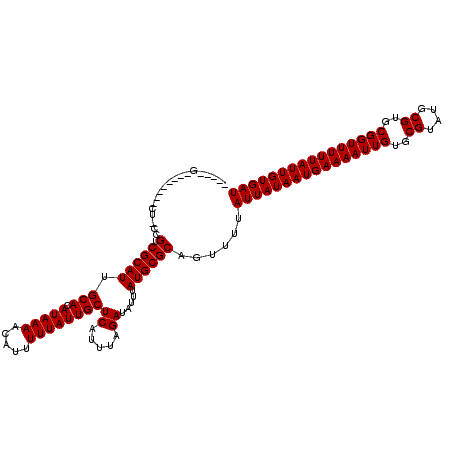
| Location | 18,298,666 – 18,298,785 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -25.85 |
| Energy contribution | -25.85 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
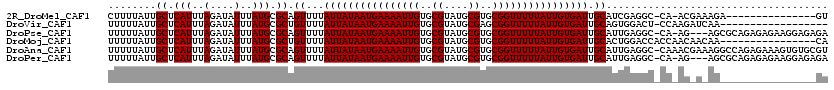
>2R_DroMel_CAF1 18298666 119 + 20766785 AUGUCGUAGAAUCGUCGCCGUCGCAUUGCACAUAAAACACUUUUAUUGCUCAUUUAGAUAUUUAUGCGCAGUUUUAUUAUAAUGAAAAUUGUGCGUAUGCGUGCGGUUUUUAUUGUGAU .((((((((((((...((....))...(((.(((((.....)))))))).......))).)))))).))).....((((((((((((((((..((....))..)))))))))))))))) ( -29.90) >DroVir_CAF1 151928 105 + 1 -----G--------CU-GCGUCGCAUUGCACAUAAAACAUUUUUAUUGCUCAUUUAGAUAUUUAUGCGCUGUUUUAUUAUAAUGAAAAUUGUGCGUAUGCGAGCGGUUUUUAUUGUGAU -----(--------((-((.(((((((((((((((((((........((.(((..(....)..))).))))))))))...(((....)))))))).)))))))))))............ ( -27.20) >DroPse_CAF1 96969 111 + 1 AUGUCG--------UUGCCGUCGCAUUGCACAUAAAACAUUUUUAUUGCUCAUUUAGAUAUUUAUGCGCAGUUUUAUUAUAAUGAAAAUUGUGCGUAUGCGUGCGGUUUUUAUUGUGAU ((((((--------.(((....))).)).))))...........(((((.(((..(....)..))).)))))...((((((((((((((((..((....))..)))))))))))))))) ( -29.30) >DroGri_CAF1 155834 105 + 1 -----G--------CU-GCGUCGCAUUGCACAUAAAACAUUUUUAUUGCUCAUUUAGAUAUUUAUGCGCUGUUUUAUUAUAAUGAAAAUUGUGCGUAUGCGUGCGGUUUUUAUUGUGAU -----.--------..-(((.(((((.(((.(((((.....))))))))((.....)).....))))).)))...((((((((((((((((..((....))..)))))))))))))))) ( -27.70) >DroMoj_CAF1 167835 105 + 1 -----G--------CU-GCGUCGCAUUGCACAUAAAACAUUUUUAUUGCUCAUUUAGAUAUUUAUGCGCUGUUUUAUUAUAAUGAAAAUUGUGCGUAUGCGUGCGGUUUUUAUUGUGAU -----.--------..-(((.(((((.(((.(((((.....))))))))((.....)).....))))).)))...((((((((((((((((..((....))..)))))))))))))))) ( -27.70) >DroPer_CAF1 97189 111 + 1 AUGUCG--------UUGCCGUCGCAUUGCACAUAAAACAUUUUUAUUGCUCAUUUAGAUAUUUAUGCGCAGUUUUAUUAUAAUGAAAAUUGUGCGUAUGCGUGCGGUUUUUAUUGUGAU ((((((--------.(((....))).)).))))...........(((((.(((..(....)..))).)))))...((((((((((((((((..((....))..)))))))))))))))) ( -29.30) >consensus _____G________CU_CCGUCGCAUUGCACAUAAAACAUUUUUAUUGCUCAUUUAGAUAUUUAUGCGCAGUUUUAUUAUAAUGAAAAUUGUGCGUAUGCGUGCGGUUUUUAUUGUGAU ...................(.(((((.(((.(((((.....))))))))((.....)).....))))))......((((((((((((((((..((....))..)))))))))))))))) (-25.85 = -25.85 + -0.00)

| Location | 18,298,666 – 18,298,785 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -18.71 |
| Consensus MFE | -15.05 |
| Energy contribution | -15.22 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18298666 119 - 20766785 AUCACAAUAAAAACCGCACGCAUACGCACAAUUUUCAUUAUAAUAAAACUGCGCAUAAAUAUCUAAAUGAGCAAUAAAAGUGUUUUAUGUGCAAUGCGACGGCGACGAUUCUACGACAU .((...........((((.((....))......................((((((((((...((..............))...)))))))))).)))).((....)).......))... ( -19.24) >DroVir_CAF1 151928 105 - 1 AUCACAAUAAAAACCGCUCGCAUACGCACAAUUUUCAUUAUAAUAAAACAGCGCAUAAAUAUCUAAAUGAGCAAUAAAAAUGUUUUAUGUGCAAUGCGACGC-AG--------C----- ...............((((((((..(((((.............(((((((...........((.....))..........)))))))))))).)))))).))-..--------.----- ( -19.81) >DroPse_CAF1 96969 111 - 1 AUCACAAUAAAAACCGCACGCAUACGCACAAUUUUCAUUAUAAUAAAACUGCGCAUAAAUAUCUAAAUGAGCAAUAAAAAUGUUUUAUGUGCAAUGCGACGGCAA--------CGACAU ..............((((.((....))......................((((((((((..((.....))(((.......))))))))))))).)))).((....--------)).... ( -19.70) >DroGri_CAF1 155834 105 - 1 AUCACAAUAAAAACCGCACGCAUACGCACAAUUUUCAUUAUAAUAAAACAGCGCAUAAAUAUCUAAAUGAGCAAUAAAAAUGUUUUAUGUGCAAUGCGACGC-AG--------C----- ...............((.(((((..(((((.............(((((((...........((.....))..........)))))))))))).)))))..))-..--------.----- ( -16.91) >DroMoj_CAF1 167835 105 - 1 AUCACAAUAAAAACCGCACGCAUACGCACAAUUUUCAUUAUAAUAAAACAGCGCAUAAAUAUCUAAAUGAGCAAUAAAAAUGUUUUAUGUGCAAUGCGACGC-AG--------C----- ...............((.(((((..(((((.............(((((((...........((.....))..........)))))))))))).)))))..))-..--------.----- ( -16.91) >DroPer_CAF1 97189 111 - 1 AUCACAAUAAAAACCGCACGCAUACGCACAAUUUUCAUUAUAAUAAAACUGCGCAUAAAUAUCUAAAUGAGCAAUAAAAAUGUUUUAUGUGCAAUGCGACGGCAA--------CGACAU ..............((((.((....))......................((((((((((..((.....))(((.......))))))))))))).)))).((....--------)).... ( -19.70) >consensus AUCACAAUAAAAACCGCACGCAUACGCACAAUUUUCAUUAUAAUAAAACAGCGCAUAAAUAUCUAAAUGAGCAAUAAAAAUGUUUUAUGUGCAAUGCGACGC_AA________C_____ ..............((((.((....)).......................(((((((((..((.....))(((.......))))))))))))..))))..................... (-15.05 = -15.22 + 0.17)

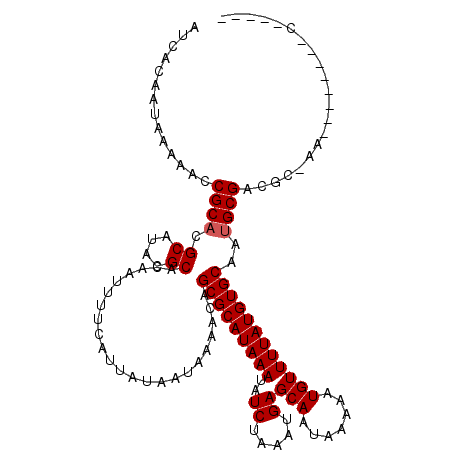
| Location | 18,298,705 – 18,298,808 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -23.57 |
| Energy contribution | -23.57 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
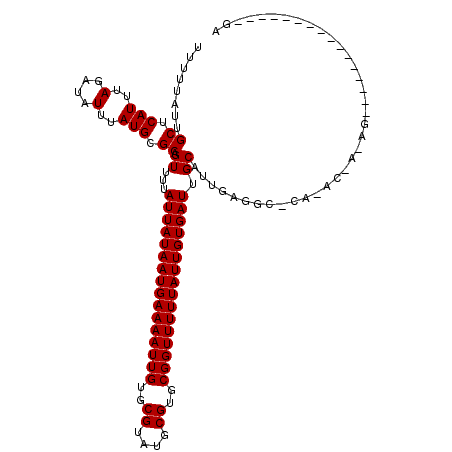
>2R_DroMel_CAF1 18298705 103 + 20766785 CUUUUAUUGCUCAUUUAGAUAUUUAUGCGCAGUUUUAUUAUAAUGAAAAUUGUGCGUAUGCGUGCGGUUUUUAUUGUGAUUGCAUCGAGGC-CA-ACGAAAGA---------------GU (((((..(((.(((..(....)..))).)))((...((((((((((((((((..((....))..)))))))))))))))).))........-..-..))))).---------------.. ( -27.50) >DroVir_CAF1 151953 101 + 1 UUUUUAUUGCUCAUUUAGAUAUUUAUGCGCUGUUUUAUUAUAAUGAAAAUUGUGCGUAUGCGAGCGGUUUUUAUUGUGAUUGCAGUGGACU-CCAAGAUCAA------------------ .................(((.....(.((((((...(((((((((((((((((.((....)).))))))))))))))))).)))))).)..-.....)))..------------------ ( -26.80) >DroPse_CAF1 97000 115 + 1 UUUUUAUUGCUCAUUUAGAUAUUUAUGCGCAGUUUUAUUAUAAUGAAAAUUGUGCGUAUGCGUGCGGUUUUUAUUGUGAUUGCAUUGAGGC-CA-AG---AGCGCAGAGAGAAGGAGAGA .........(((.(((.....(((.(((((.((...((((((((((((((((..((....))..)))))))))))))))).)).(((....-))-).---.))))).))).))))))... ( -32.40) >DroMoj_CAF1 167860 104 + 1 UUUUUAUUGCUCAUUUAGAUAUUUAUGCGCUGUUUUAUUAUAAUGAAAAUUGUGCGUAUGCGUGCGGUUUUUAUUGUGAUUGCACUGGACCACCAACAACAA----------------CA ........((.(((..(....)..))).))(((...((((((((((((((((..((....))..)))))))))))))))).))).(((....))).......----------------.. ( -25.50) >DroAna_CAF1 101098 119 + 1 UUUUUAUUGCUCAUUUAGAUAUUUAUGCGCAGUUUUAUUAUAAUGAAAAUUGUGCGUAUGCGUGCGGUUUUUAUUGUGAUUGCAUUGAGGC-CAAACGAAAGGCCAGAGAAAGUGUGCGU ........................(((((((.((((((((((((((((((((..((....))..))))))))))))))))........(((-(........))))...)))).))))))) ( -38.90) >DroPer_CAF1 97220 115 + 1 UUUUUAUUGCUCAUUUAGAUAUUUAUGCGCAGUUUUAUUAUAAUGAAAAUUGUGCGUAUGCGUGCGGUUUUUAUUGUGAUUGCAUUGAGGC-CA-AG---AGCGCAGAGAGAAGGAGAGA .........(((.(((.....(((.(((((.((...((((((((((((((((..((....))..)))))))))))))))).)).(((....-))-).---.))))).))).))))))... ( -32.40) >consensus UUUUUAUUGCUCAUUUAGAUAUUUAUGCGCAGUUUUAUUAUAAUGAAAAUUGUGCGUAUGCGUGCGGUUUUUAUUGUGAUUGCAUUGAGGC_CA_AC_A_AG________________GA ........((.(((..(....)..))).)).((...((((((((((((((((..((....))..)))))))))))))))).))..................................... (-23.57 = -23.57 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:04 2006