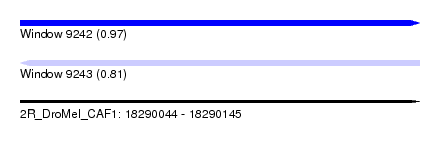
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,290,044 – 18,290,145 |
| Length | 101 |
| Max. P | 0.969746 |

| Location | 18,290,044 – 18,290,145 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.74 |
| Mean single sequence MFE | -24.09 |
| Consensus MFE | -16.89 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18290044 101 + 20766785 CAGCGAAACACAGAACUUAUCCUGGCCAAAAGCAUCCUUGCCAUAUACA--UAUGCAC---------------CAAGGAUGCUCCGCCAGCCACACAUACAUAUGCGAUAGCAUCACA ..(.((...............(((((....((((((((((.((((....--))))...---------------))))))))))..))))).............(((....))))).). ( -27.10) >DroSec_CAF1 87340 101 + 1 CAGCGAAACACAGAACUUAUCCUGGCCAAAAGCAUCCUUGUCAUAUACA--UAUGCAC---------------CAAGGAUGCUCCGCCGGCCACACAUACAUAUGCAAUAGCAUCACA ..(.((................(((((...((((((((((.((((....--))))...---------------)))))))))).....)))))..........(((....))))).). ( -26.70) >DroSim_CAF1 88574 101 + 1 CAGCGAAACACAGAACUUAUCCUGGCCAAAAGCAUUCUUGUCAUAUACA--UAUGCAC---------------CAAGGAUGCUCCGCCAGCCACACAUACAUAUGCGAUAUCAUCACA ............((...(((((((((....((((((((((.((((....--))))...---------------))))))))))..))))).....(((....))).))))...))... ( -22.90) >DroEre_CAF1 91002 112 + 1 CAACGAAACACAGAACUUAUCCUGGCCAAAAGCAUCCUUGUCAUAUACA--UAUGCACCGAAAGUUAAGGCAGCAAGUAUGCUCCGGCAGCCACAC----AUAUGCGAUAGUAUCACA ....((.((.(((........)))(((...(((((.(((((........--..(((..(....).....)))))))).)))))..)))........----..........)).))... ( -21.30) >DroYak_CAF1 88611 113 + 1 CAGCGAAACACAGAAUUUAUCCUGGCCAAAAGCAUCCUUGUCAUAUACACACAAGCACCGAAAGUUAAGGCAGCAAGGAUGCUCU-GCAGCCACAC----AUAUGCGAUAGUAUCACA ............((..((((((((((..(.(((((((((((.............((..(....).....)).))))))))))).)-...))))...----....).)))))..))... ( -22.45) >consensus CAGCGAAACACAGAACUUAUCCUGGCCAAAAGCAUCCUUGUCAUAUACA__UAUGCAC_______________CAAGGAUGCUCCGCCAGCCACACAUACAUAUGCGAUAGCAUCACA ......................((((....((((((((((.................................))))))))))......)))).........((((....)))).... (-16.89 = -16.93 + 0.04)

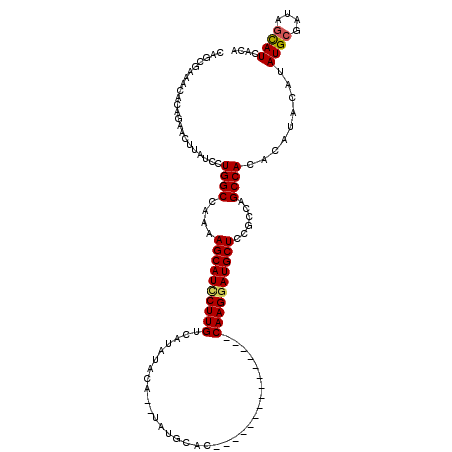
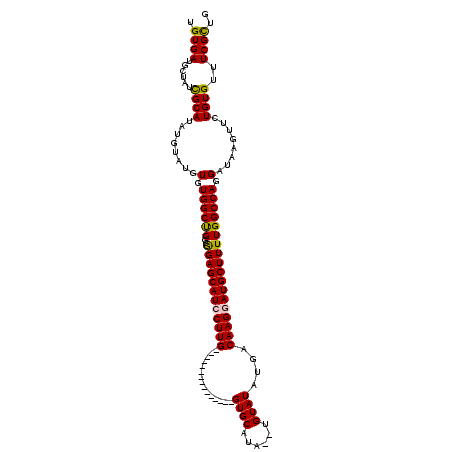
| Location | 18,290,044 – 18,290,145 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.74 |
| Mean single sequence MFE | -32.53 |
| Consensus MFE | -27.44 |
| Energy contribution | -27.20 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18290044 101 - 20766785 UGUGAUGCUAUCGCAUAUGUAUGUGUGGCUGGCGGAGCAUCCUUG---------------GUGCAUA--UGUAUAUGGCAAGGAUGCUUUUGGCCAGGAUAAGUUCUGUGUUUCGCUG .((((.((..(((((((....)))))))(((((((((((((((((---------------((((...--.))))....))))))))))))..)))))............)).)))).. ( -37.20) >DroSec_CAF1 87340 101 - 1 UGUGAUGCUAUUGCAUAUGUAUGUGUGGCCGGCGGAGCAUCCUUG---------------GUGCAUA--UGUAUAUGACAAGGAUGCUUUUGGCCAGGAUAAGUUCUGUGUUUCGCUG .((((.((....(((((....)))))((((((..(((((((((((---------------...((((--....)))).))))))))))))))))).(((.....)))..)).)))).. ( -36.40) >DroSim_CAF1 88574 101 - 1 UGUGAUGAUAUCGCAUAUGUAUGUGUGGCUGGCGGAGCAUCCUUG---------------GUGCAUA--UGUAUAUGACAAGAAUGCUUUUGGCCAGGAUAAGUUCUGUGUUUCGCUG .((((.(((((((((((....)))))).((((((((((((.((((---------------...((((--....)))).)))).)))))))..)))))..........))))))))).. ( -31.40) >DroEre_CAF1 91002 112 - 1 UGUGAUACUAUCGCAUAU----GUGUGGCUGCCGGAGCAUACUUGCUGCCUUAACUUUCGGUGCAUA--UGUAUAUGACAAGGAUGCUUUUGGCCAGGAUAAGUUCUGUGUUUCGUUG .(.(((((....((.(((----.(.((((((..(((((((.((((..(((.........))).((((--....)))).)))).))))))))))))).)))).))...))))).).... ( -24.80) >DroYak_CAF1 88611 113 - 1 UGUGAUACUAUCGCAUAU----GUGUGGCUGC-AGAGCAUCCUUGCUGCCUUAACUUUCGGUGCUUGUGUGUAUAUGACAAGGAUGCUUUUGGCCAGGAUAAAUUCUGUGUUUCGCUG .((((.((..........----.(.((((((.-((((((((((((..(((.........)))((......))......)))))))))))))))))).)...........)).)))).. ( -32.85) >consensus UGUGAUGCUAUCGCAUAUGUAUGUGUGGCUGGCGGAGCAUCCUUG_______________GUGCAUA__UGUAUAUGACAAGGAUGCUUUUGGCCAGGAUAAGUUCUGUGUUUCGCUG .((((......((((........(.((((((..((((((((((((...............((((......))))....)))))))))))))))))).)........))))..)))).. (-27.44 = -27.20 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:58 2006