
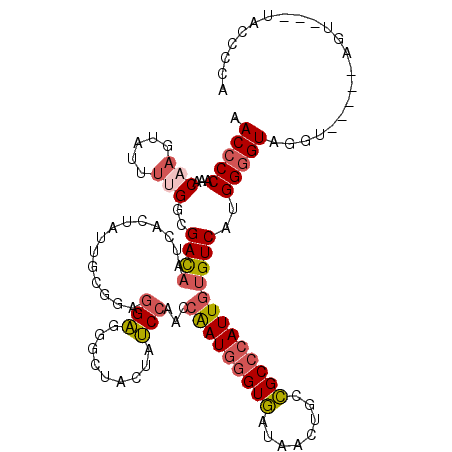
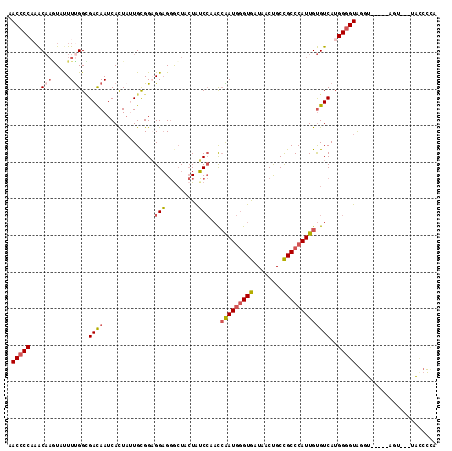
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,271,534 – 18,271,669 |
| Length | 135 |
| Max. P | 0.981606 |

| Location | 18,271,534 – 18,271,643 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.57 |
| Mean single sequence MFE | -36.29 |
| Consensus MFE | -21.81 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18271534 109 + 20766785 AACCCCAAACAAGUAUUUUGGUGACAAUCACUAUUGCGGAGGAGGGCUACUGUCCAAUCAAUGGGUGAUUACUGCCGCCCAUUGUGUCAUGGGGUAGGU-----AGU---UACCUCA .((((((.....(((...(((((.....))))).))).((.(.((((....))))...(((((((((........)))))))))).)).))))))((((-----...---.)))).. ( -38.90) >DroVir_CAF1 110876 111 + 1 UACACCUCGCCUUUACUUUGGGGAUAAUCAUUUUUGUGGUGGUGCGAUAAUAGCACCCAGAUAUGUAUUAACUGCUGCCCAUUGCGUCAUGGAGUGGGU-----AGAAAU-GCAUUU ......((.((........)).))..(((((....)))))(((((.......))))).....(((((((.....(((((((((.(.....).)))))))-----)).)))-)))).. ( -31.90) >DroSec_CAF1 68468 109 + 1 AACCCCAAACAAAUUUUUUGGCGACAAUCACUACUGCGGAGGAGGGCUACUAUCCAACCAAUGGGUGAUAACUGCCGCCCAUUGUGUCAUGGGGUAGGU-----AAU---UACCUCA .((((((..(((.....)))..((((...........(((..((.....)).)))...(((((((((........))))))))))))).))))))((((-----...---.)))).. ( -35.30) >DroSim_CAF1 70247 109 + 1 AACCCCAAACAAAUUCUUUGGCGACAAUCACUACUGCGGAGGAGGGCUACUAUCCAACCAAUGGGUGAUAACUGCCGCCCAUUGUGUCAUGGGGUAGGU-----AGU---UACCCCA ....(((((.......))))).((((...........(((..((.....)).)))...(((((((((........))))))))))))).((((((((..-----..)---))))))) ( -35.60) >DroEre_CAF1 70807 109 + 1 AACCCCCAACAAGUAUUUUGGCGACAAUCACUAUUGCGGAGGAGGACUUCUAUCCAACCAAUGGGUGCUUACCGCCGCCCAUUGUGUCAUGGGGUAGGU-----UGU---UGUCCCA ..(((((((..(((...(((....)))..))).))).)).)).((((.(((((((.((((((((((((.....).))))))))).))....))))))).-----...---.)))).. ( -36.80) >DroYak_CAF1 70198 114 + 1 AACCCCAAACAAGUAUUUCGGCGACAAUCACUAUUGCGGUGGAGGGCUUCUAUCCAACCGAUGGGUGCUCACUGCCGCCCAUUGUGUCAUGGGGUAGGUACUAUAGU---UAACCAA .((((((.....((......))((((.........((((..(.((((.((((((.....)))))).)))).)..))))......)))).)))))).(((((....))---..))).. ( -39.26) >consensus AACCCCAAACAAGUAUUUUGGCGACAAUCACUAUUGCGGAGGAGGGCUACUAUCCAACCAAUGGGUGAUAACUGCCGCCCAUUGUGUCAUGGGGUAGGU_____AGU___UACCCCA .(((((...(((.....)))..((((..............(((.........)))...(((((((((........)))))))))))))..)))))...................... (-21.81 = -22.40 + 0.59)

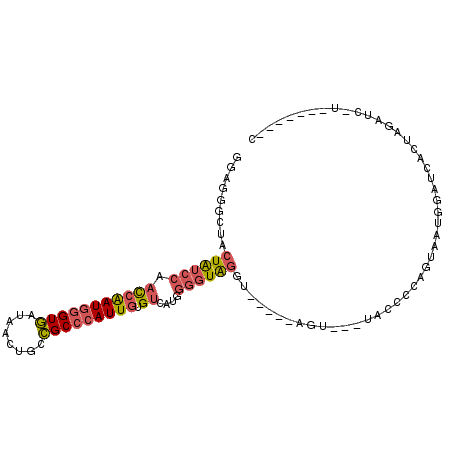
| Location | 18,271,574 – 18,271,669 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 70.78 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -15.90 |
| Energy contribution | -16.32 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18271574 95 + 20766785 GGAGGGCUACUGUCCAAUCAAUGGGUGAUUACUGCCGCCCAUUGUGUCAUGGGGUAGGU-----AGU---UACCUCAGUAAUGGAUCACUGGAUCUUAAU--AUC .((((((((((.((((..(((((((((........))))))))).....))))...)))-----)))---..))))......(((((....)))))....--... ( -32.80) >DroVir_CAF1 110916 94 + 1 GGUGCGAUAAUAGCACCCAGAUAUGUAUUAACUGCUGCCCAUUGCGUCAUGGAGUGGGU-----AGAAAU-GCAUUUGUAG---ACCGCUAGAUCAAAUUUA--A (((((.......)))))..((((((((((.....(((((((((.(.....).)))))))-----)).)))-))))...(((---....))).))).......--. ( -26.80) >DroSec_CAF1 68508 89 + 1 GGAGGGCUACUAUCCAACCAAUGGGUGAUAACUGCCGCCCAUUGUGUCAUGGGGUAGGU-----AAU---UACCUCAGUAAUCGGUAACUGGAUCAU-------- .((((....((((((.(((((((((((........))))))))).))....))))))..-----...---..)))).......(((......)))..-------- ( -28.70) >DroSim_CAF1 70287 89 + 1 GGAGGGCUACUAUCCAACCAAUGGGUGAUAACUGCCGCCCAUUGUGUCAUGGGGUAGGU-----AGU---UACCCCAGUAAUCGGUCACUGGAUCUU-------- ((..(((((((((((.(((((((((((........))))))))).))....))))).))-----)))---).))(((((........))))).....-------- ( -33.30) >DroEre_CAF1 70847 89 + 1 GGAGGACUUCUAUCCAACCAAUGGGUGCUUACCGCCGCCCAUUGUGUCAUGGGGUAGGU-----UGU---UGUCCCAGUAAUGGAUCUUCA-CUU-------AAC (((((((.(((((((.((((((((((((.....).))))))))).))....))))))).-----.))---)...(((....)))..)))).-...-------... ( -31.30) >DroYak_CAF1 70238 92 + 1 GGAGGGCUUCUAUCCAACCGAUGGGUGCUCACUGCCGCCCAUUGUGUCAUGGGGUAGGUACUAUAGU---UAACCAAGUAAUGGAUCUCUA-AUC---------C ((((((..(((((((.((((((((((((.....).))))))))).))....)))))))..)).....---...(((.....)))..)))).-...---------. ( -29.50) >consensus GGAGGGCUACUAUCCAACCAAUGGGUGAUAACUGCCGCCCAUUGUGUCAUGGGGUAGGU_____AGU___UACCCCAGUAAUGGAUCACUAGAUC_U_______C .........((((((.(((((((((((........))))))))).))....))))))................................................ (-15.90 = -16.32 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:52 2006