| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,210,299 – 18,210,418 |
| Length | 119 |
| Max. P | 0.825395 |

| Location | 18,210,299 – 18,210,418 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.77 |
| Mean single sequence MFE | -39.82 |
| Consensus MFE | -21.04 |
| Energy contribution | -22.43 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.53 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18210299 119 + 20766785 UGAAACAGUUGAUUUUCCUGCUGAUUUGUUUGAGCUGCGGCACCUGCUCCAUUUACGCACUGAAAUGCAGAUCCCAGGAGGGACUAAGUGAAGCGGAGCUCAAGCGAACUGUGCGCAAC ((..((((((((((........))))(((((((((.((((...))))(((.((((((((......)))((.((((....))))))..)))))..))))))))))))))))))...)).. ( -36.40) >DroSec_CAF1 8165 119 + 1 UGAAACAGUUGAUUUUGCUGUUUAUUUGCUUGAGCUGCGGCACCUGCUCCAUUUUCGCAUUGAAAUGCAGAUCCCAGGAAGGACUAAGUGAAGCGGAGCUCAAGCGAACUGUGCGCAAC ..(((((((.......))))))).((((((((((((.((.(((.((.(((......((((....))))...((....)).))).)).)))...)).))))))))))))........... ( -39.50) >DroSim_CAF1 8165 119 + 1 UGAAACAGUUGAUUCUGCUGCUGAUUUGCUUGAGCAGCGGCACCUGCUCCAUUUACGCACUGAAAUGCAGAUCCCAGGAAGGACUCAGUGAAGCGGAGCUCAAGCGAACUGUGCGCAAC ((..((((((.....(((((((.(......).)))))))((....(((((.((((((((......))).(((((......))).)).)))))..)))))....)).))))))...)).. ( -39.20) >DroEre_CAF1 8148 116 + 1 UGAA---UUUAAUUUUGCUGUUGAUUGGCUUGAGCUGCGGCAUCUGCUCCAUUUACGCACUGAAAUGCAGAUCCCAGGAAGGAUUAGGUGAAGCGGAGCUCAAGCGAACUGUGCGCAGC ....---.........((((((..((.(((((((((.((.((((((.(((......(((......)))...((....)).))).))))))...)).))))))))).))..)...))))) ( -39.00) >DroYak_CAF1 8330 107 + 1 UGAAACAGUUGAUUCUGUUGCUGCUUUGCCUGAGCUG------------CAUUUACGCACUGAAGUGCAGAUCCCAGGAAGGAUUAAGUGAAGCGGAGCUCAAGCGAACUGUGCGCAGU ...(((((......)))))((((((((((.((((((.------------(.(((((((((....)))).(((((......)))))..)))))..).)))))).)))))......))))) ( -39.10) >DroMoj_CAF1 32060 110 + 1 UGCGCCUGCUGCUGUUGCUUUUACUCAGCUUUAGCUGCAGC---------AUCAACUGCCUCAAGUGUCGCUCGCAGGAUGGCGCCGGCGAUGCGGAGCUGAAGCGGAUUGUGCGCACU (((((((((.((((..(((.......)))..)))).)))).---------..(((((((.(((.((..(((((((.((......)).)))).)))..))))).)))).))).))))).. ( -45.70) >consensus UGAAACAGUUGAUUUUGCUGCUGAUUUGCUUGAGCUGCGGCACCUGCUCCAUUUACGCACUGAAAUGCAGAUCCCAGGAAGGACUAAGUGAAGCGGAGCUCAAGCGAACUGUGCGCAAC ..............((((.((...((((((((((((.((.................(((......)))...(((......)))..........)).))))))))))))....)))))). (-21.04 = -22.43 + 1.39)

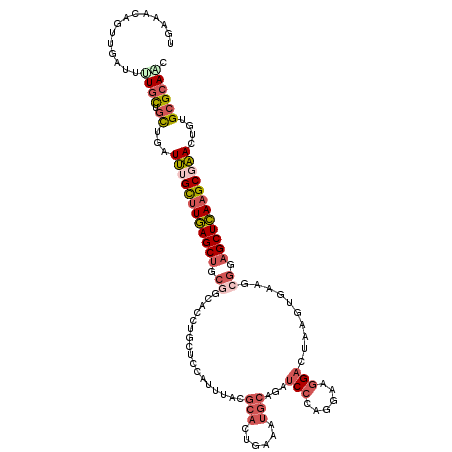
| Location | 18,210,299 – 18,210,418 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.77 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -21.10 |
| Energy contribution | -22.30 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18210299 119 - 20766785 GUUGCGCACAGUUCGCUUGAGCUCCGCUUCACUUAGUCCCUCCUGGGAUCUGCAUUUCAGUGCGUAAAUGGAGCAGGUGCCGCAGCUCAAACAAAUCAGCAGGAAAAUCAACUGUUUCA ((((((((((((((....)))))..((((((.((((((((....)))))..((((....)))).))).))))))..))).))))))...........(((((.........)))))... ( -38.50) >DroSec_CAF1 8165 119 - 1 GUUGCGCACAGUUCGCUUGAGCUCCGCUUCACUUAGUCCUUCCUGGGAUCUGCAUUUCAAUGCGAAAAUGGAGCAGGUGCCGCAGCUCAAGCAAAUAAACAGCAAAAUCAACUGUUUCA .((((.....(((.(((((((((.(((((((....(((((....))))).(((((....)))))....)))))).(....)).))))))))).))).....)))).............. ( -35.50) >DroSim_CAF1 8165 119 - 1 GUUGCGCACAGUUCGCUUGAGCUCCGCUUCACUGAGUCCUUCCUGGGAUCUGCAUUUCAGUGCGUAAAUGGAGCAGGUGCCGCUGCUCAAGCAAAUCAGCAGCAGAAUCAACUGUUUCA .(((((((((((((....)))))..((......((.((((....)))))).))......)))))))).((((((((.((...(((((...((......)))))))...)).)))))))) ( -37.70) >DroEre_CAF1 8148 116 - 1 GCUGCGCACAGUUCGCUUGAGCUCCGCUUCACCUAAUCCUUCCUGGGAUCUGCAUUUCAGUGCGUAAAUGGAGCAGAUGCCGCAGCUCAAGCCAAUCAACAGCAAAAUUAAA---UUCA ((((......(((.(((((((((.(((((((....(((((....)))))..((((....)))).....)))))).(....)).))))))))).)))...)))).........---.... ( -34.20) >DroYak_CAF1 8330 107 - 1 ACUGCGCACAGUUCGCUUGAGCUCCGCUUCACUUAAUCCUUCCUGGGAUCUGCACUUCAGUGCGUAAAUG------------CAGCUCAGGCAAAGCAGCAACAGAAUCAACUGUUUCA .((((......((.(((((((((..((((.((...(((((....)))))..((((....)))))).)).)------------)))))))))))).)))).(((((......)))))... ( -31.80) >DroMoj_CAF1 32060 110 - 1 AGUGCGCACAAUCCGCUUCAGCUCCGCAUCGCCGGCGCCAUCCUGCGAGCGACACUUGAGGCAGUUGAU---------GCUGCAGCUAAAGCUGAGUAAAAGCAACAGCAGCAGGCGCA ..(((((.......(((((((...(((.((((.((......)).)))))))....))))))).((((.(---------(((.((((....))))......)))).)))).....))))) ( -42.30) >consensus GUUGCGCACAGUUCGCUUGAGCUCCGCUUCACUUAGUCCUUCCUGGGAUCUGCAUUUCAGUGCGUAAAUGGAGCAGGUGCCGCAGCUCAAGCAAAUCAACAGCAAAAUCAACUGUUUCA .((((.....(((.(((((((((.((.........(((((....)))))..((((....)))).................)).))))))))).))).....)))).............. (-21.10 = -22.30 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:33 2006