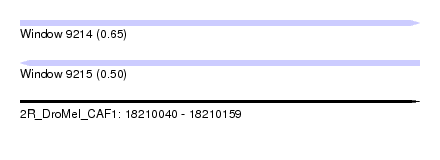
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,210,040 – 18,210,159 |
| Length | 119 |
| Max. P | 0.646463 |

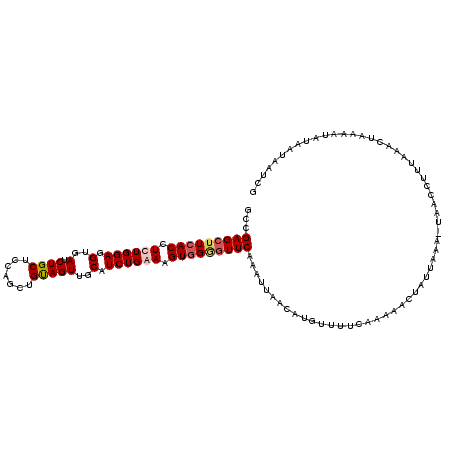
| Location | 18,210,040 – 18,210,159 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.50 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -21.74 |
| Energy contribution | -22.68 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18210040 119 + 20766785 AUCGAGCUUCACCUCUGGAGGUGAUCUGCUCCAGCUGUAGUUGCAUCUGGAAGUGGGGUUCAAUUUAACAUGUUUUCAUAAACUUUUAAAAUAACCUUUAUUGUAAAAUAUAAUCAUCG ...(((((((((.((..((.(..(.((((.......)))))..).))..)).)))))))))........(((((((((((((..((......))..)))).)).)))))))........ ( -28.60) >DroSec_CAF1 7909 118 + 1 GCCGAGCUUCACCUCUGGAGGUGAUCUGCUCCAGCUGUAGUUGCAUCUGGAUGUGGAGUUCAGAUUAACAUGUUUUCAAAAACUAUUAAA-AGCCCUUUAAACUAAAAUAUAAUAAUCA ...(((((((((.((..((.(..(.((((.......)))))..).))..)).))))))))).(((((..(((((((..............-.............)))))))..))))). ( -30.93) >DroSim_CAF1 7907 118 + 1 GCCGAGCUUCACCUCUGGAGGUGAUCUGCUCCAGCUGUAGUUGCAUCUGAAAGUGGGGUUCAGAUUAACAUGUUUUCAAAAACUAUUAAA-UAACCUUUAAACUAAAAUAUAAAAAUCG ((.((((.((((((....))))))...))))..))....((((.(((((((.......))))))))))).....................-............................ ( -24.90) >DroEre_CAF1 7890 118 + 1 GUCGAGCUUCACCUCUGGAGGUGAUCUGCUCCAGCUGCAGUUGCAUCUGAAAGUUGCAUUCAAAUGAACAUGUUUUUAACAGCUAUUAAA-UAACCUUUAUAUUAAAAAAUAAUCUUUG ...((((.((((((....))))))...)))).((((((((......)))...(((.(((....))))))..........)))))......-............................ ( -24.30) >consensus GCCGAGCUUCACCUCUGGAGGUGAUCUGCUCCAGCUGUAGUUGCAUCUGAAAGUGGGGUUCAAAUUAACAUGUUUUCAAAAACUAUUAAA_UAACCUUUAAACUAAAAUAUAAUAAUCG ...(((((((((.((((((.(..(.((((.......)))))..).)))))).))))))))).......................................................... (-21.74 = -22.68 + 0.94)

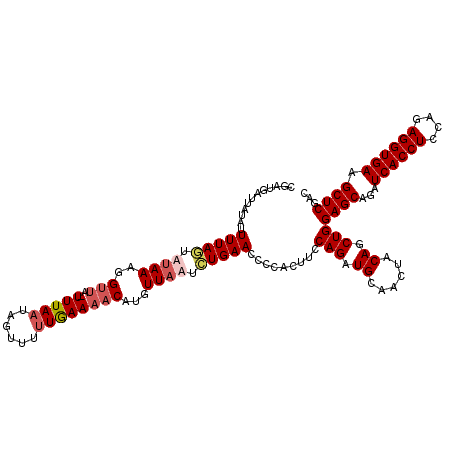
| Location | 18,210,040 – 18,210,159 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.50 |
| Mean single sequence MFE | -27.13 |
| Consensus MFE | -17.86 |
| Energy contribution | -19.18 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18210040 119 - 20766785 CGAUGAUUAUAUUUUACAAUAAAGGUUAUUUUAAAAGUUUAUGAAAACAUGUUAAAUUGAACCCCACUUCCAGAUGCAACUACAGCUGGAGCAGAUCACCUCCAGAGGUGAAGCUCGAU (((....................((((..(((((..((((....))))...)))))...)))).....(((((.((......)).)))))((...((((((....)))))).))))).. ( -23.80) >DroSec_CAF1 7909 118 - 1 UGAUUAUUAUAUUUUAGUUUAAAGGGCU-UUUAAUAGUUUUUGAAAACAUGUUAAUCUGAACUCCACAUCCAGAUGCAACUACAGCUGGAGCAGAUCACCUCCAGAGGUGAAGCUCGGC .......................(((((-.((((((((((....)))).))))))((((.........(((((.((......)).))))).))))((((((....)))))))))))... ( -26.60) >DroSim_CAF1 7907 118 - 1 CGAUUUUUAUAUUUUAGUUUAAAGGUUA-UUUAAUAGUUUUUGAAAACAUGUUAAUCUGAACCCCACUUUCAGAUGCAACUACAGCUGGAGCAGAUCACCUCCAGAGGUGAAGCUCGGC .................((((((..(((-(...))))..)))))).....(((.(((((((.......)))))))..)))....((((.(((...((((((....)))))).))))))) ( -26.40) >DroEre_CAF1 7890 118 - 1 CAAAGAUUAUUUUUUAAUAUAAAGGUUA-UUUAAUAGCUGUUAAAAACAUGUUCAUUUGAAUGCAACUUUCAGAUGCAACUGCAGCUGGAGCAGAUCACCUCCAGAGGUGAAGCUCGAC ...((((.((((((......)))))).)-)))..(((((((.........(((((((((((.......)))))))).))).)))))))((((...((((((....)))))).))))... ( -31.70) >consensus CGAUGAUUAUAUUUUAGUAUAAAGGUUA_UUUAAUAGUUUUUGAAAACAUGUUAAUCUGAACCCCACUUCCAGAUGCAACUACAGCUGGAGCAGAUCACCUCCAGAGGUGAAGCUCGAC ............(((((.((((..(((..(((((......))))))))...)))).))))).........(((.((......)).)))((((...((((((....)))))).))))... (-17.86 = -19.18 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:31 2006