
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
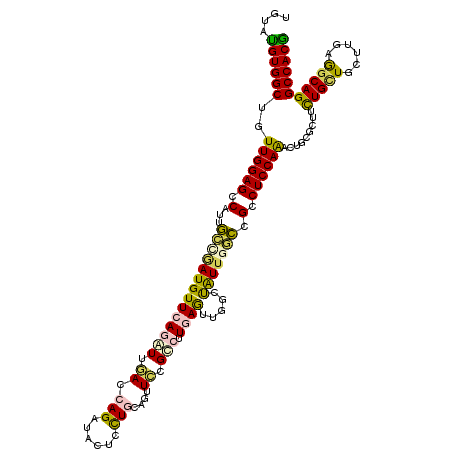
| Location | 18,209,266 – 18,209,386 |
| Length | 120 |
| Max. P | 0.921520 |

| Location | 18,209,266 – 18,209,386 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -42.19 |
| Consensus MFE | -25.68 |
| Energy contribution | -25.05 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.655237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
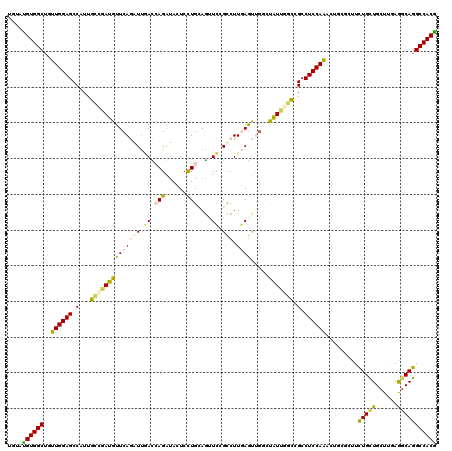
>2R_DroMel_CAF1 18209266 120 + 20766785 UGGACGUGGCUAUUGGAGCCAUUGGCAAUGUUCAAAUUUACCAGGUAGUCCUGCAGCUGCGUCUUUAGCUGGCUGUUCUCCGCCUCCAGGCGACGCUUCUGUUGCUUCAGGCAGGCCACG ....(((((((...((((....((((...............((((....))))((((((......))))))))))..))))((((..((((((((....)))))))).)))).))))))) ( -45.60) >DroVir_CAF1 34832 120 + 1 UGUAUGUGGCUGUUGGAGCCGUUGCUAAUGUUCAGGUUGAUGAGAUACUCUUGCAGUUCUGCCUUGAGUCGUGCAUUGUCUGCCUCCAGAUUGCGUUUUUGCUGCUUAAGACAGGCCACG .....(((((((((.((((.((....(((((((((((.((..(...((((..(((....)))...))))......)..)).))))...))..)))))...)).))))..)))).))))). ( -34.40) >DroPse_CAF1 7752 120 + 1 UGUAUGUGGCUGUUGGAGCCAUUUCCGAUGUUCAGGUUGACCAGGUACUCCUGGAGUUCCGCCUUGAGUUGGCUAUUGGACGCCUCCAACCUCUUCUUCUGCUGCUUGAGGCAGGCCACG .....(((((.(((((((.(...(((((((....((....(((((....)))))....))(((.......)))))))))).).)))))))........(((((......)))))))))). ( -46.50) >DroGri_CAF1 29500 120 + 1 UGUAUGUGGCAGUUGGAGCCAUUGCUGAUGUUCAAAUUGAUGAGAUACUCCUGCAAAUCGGCCUUGAGCCGUGUAUUGGCCGCCUCCAAAGUGCGUUUCUGCUGCUUCAAGCAGGCCACA ....((((((..((((((.(.((((.((..((((......))))....))..))))...((((..............))))).)))))).........(((((......))))))))))) ( -37.34) >DroMoj_CAF1 30967 120 + 1 UGGAUGUGGCUAUUGGAGCCGUUGGCUAUGUUCAGAUCAAUCAGAUAGUCCUGCAGCUCGGCCUUCAAUUUUGCAUUUUCUGCCUCCAAGUUGCGUUUCUGCUGCUUGAGGCAGGCCACG .....(((((.((((((((((..((((........(((.....)))........))))))).)))))))..........(((((((.((((.(((....))).)))))))))))))))). ( -42.79) >DroPer_CAF1 7749 120 + 1 UGUAUGUGGCUGUUGGAGCCAUUUCCGAUGUUCAGGUUGACCAGGUACUCCUGGAGUUCCGCCUUGAGUUGGCUAUUGGACGCCUCCAACCUCUUCUUCUGCUGCUUGAGGCAGGCCACG .....(((((.(((((((.(...(((((((....((....(((((....)))))....))(((.......)))))))))).).)))))))........(((((......)))))))))). ( -46.50) >consensus UGUAUGUGGCUGUUGGAGCCAUUGCCGAUGUUCAGAUUGACCAGAUACUCCUGCAGUUCCGCCUUGAGUUGGCUAUUGGCCGCCUCCAAACUGCGCUUCUGCUGCUUGAGGCAGGCCACG ....((((((..((((((.(...((((((((((((((.((.(((......)))....)).))).)))).....))))))).).)))))).........(((((......))))))))))) (-25.68 = -25.05 + -0.63)

| Location | 18,209,266 – 18,209,386 |
|---|---|
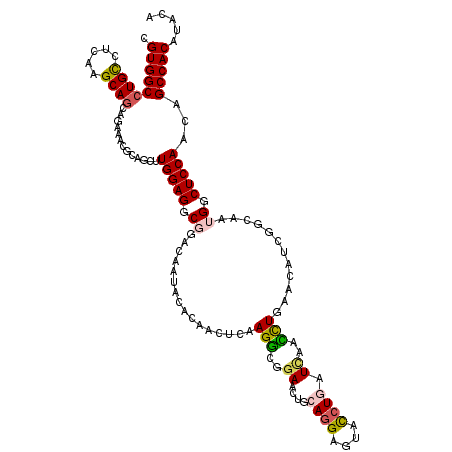
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -39.45 |
| Consensus MFE | -21.40 |
| Energy contribution | -21.46 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18209266 120 - 20766785 CGUGGCCUGCCUGAAGCAACAGAAGCGUCGCCUGGAGGCGGAGAACAGCCAGCUAAAGACGCAGCUGCAGGACUACCUGGUAAAUUUGAACAUUGCCAAUGGCUCCAAUAGCCACGUCCA ((((((..(((....(((((((..((((((((....)))((.......)).......)))))..)))((((....)))).............))))....))).......)))))).... ( -39.40) >DroVir_CAF1 34832 120 - 1 CGUGGCCUGUCUUAAGCAGCAAAAACGCAAUCUGGAGGCAGACAAUGCACGACUCAAGGCAGAACUGCAAGAGUAUCUCAUCAACCUGAACAUUAGCAACGGCUCCAACAGCCACAUACA .((((((((((((.((..((......))...)).))))))).....((.((((((...(((....)))..))))...(((......)))..........)))).......)))))..... ( -30.50) >DroPse_CAF1 7752 120 - 1 CGUGGCCUGCCUCAAGCAGCAGAAGAAGAGGUUGGAGGCGUCCAAUAGCCAACUCAAGGCGGAACUCCAGGAGUACCUGGUCAACCUGAACAUCGGAAAUGGCUCCAACAGCCACAUACA .(((((((((........))))........(((((((.(.(((....(((.......)))((....(((((....)))))....))........)))...).))))))).)))))..... ( -45.10) >DroGri_CAF1 29500 120 - 1 UGUGGCCUGCUUGAAGCAGCAGAAACGCACUUUGGAGGCGGCCAAUACACGGCUCAAGGCCGAUUUGCAGGAGUAUCUCAUCAAUUUGAACAUCAGCAAUGGCUCCAACUGCCACAUACA (((((((((((......)))))....(((..((((....((((.......)))).....))))..))).(((((...(((......)))....((....)))))))....)))))).... ( -38.10) >DroMoj_CAF1 30967 120 - 1 CGUGGCCUGCCUCAAGCAGCAGAAACGCAACUUGGAGGCAGAAAAUGCAAAAUUGAAGGCCGAGCUGCAGGACUAUCUGAUUGAUCUGAACAUAGCCAACGGCUCCAAUAGCCACAUCCA .(((((((((((((((..((......))..))).)))))))....................((((((..((.(((((.((....)).))...)))))..)))))).....)))))..... ( -38.50) >DroPer_CAF1 7749 120 - 1 CGUGGCCUGCCUCAAGCAGCAGAAGAAGAGGUUGGAGGCGUCCAAUAGCCAACUCAAGGCGGAACUCCAGGAGUACCUGGUCAACCUGAACAUCGGAAAUGGCUCCAACAGCCACAUACA .(((((((((........))))........(((((((.(.(((....(((.......)))((....(((((....)))))....))........)))...).))))))).)))))..... ( -45.10) >consensus CGUGGCCUGCCUCAAGCAGCAGAAACGCAGCUUGGAGGCGGACAAUACACAACUCAAGGCGGAACUGCAGGAGUACCUGAUCAACCUGAACAUCGGCAAUGGCUCCAACAGCCACAUACA .(((((((((.....)))).............(((((.((................(((..((....((((....)))).))..)))............)).)))))...)))))..... (-21.40 = -21.46 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:29 2006