
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,207,181 – 18,207,274 |
| Length | 93 |
| Max. P | 0.713213 |

| Location | 18,207,181 – 18,207,274 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.93 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -17.00 |
| Energy contribution | -16.95 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
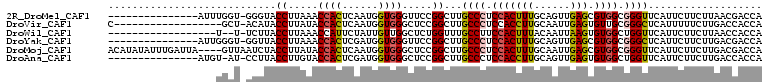
>2R_DroMel_CAF1 18207181 93 + 20766785 ---------------AUUUGGU-GGGUACCUUAAACCACUCAAUGGUGGGUUCCGGCUUGCCCUCCACUUUGCAGUUGAGCGUGGCGGGUUCAUUCUUCUUAACGACCA ---------------....(((-.((.((((...((((.....)))))))).)).))).((((.((((...((......)))))).))))................... ( -25.90) >DroVir_CAF1 32052 90 + 1 C------------------GCU-ACAUACCUUAUACCACUCAAUGGUGGGCUCCGGCUUGCCCUCCACCUUGCAAUUGAGUGUUGCGGGCUCAUUUUUCUUGACCACCA (------------------((.-.............((((((((((.((((........)))).)))........)))))))..)))((.(((.......))))).... ( -23.09) >DroWil_CAF1 4872 88 + 1 ------------------U--U-UCUUACCUUAAACCAUUCUAUUGUUGGCUCUGGUUUGCCUUCCACUUUACAAUUAAGUGUGGCUGGUUCAUUCUUCUUAACCACCA ------------------.--.-.........((((((..(((....)))...))))))(((...(((((.......))))).)))(((((..........)))))... ( -15.60) >DroYak_CAF1 5197 93 + 1 ---------------AUUGGGU-GGUUACCUUAAACCACUCGAUGGUGGGUUCCGGCUUGCCCUCCACUUUGCAGUUGAGCGUGGCGGGCUCAUUCUUCUUGACGACCA ---------------(((((((-((((......)))))))))))(((.(..........((((.((((...((......)))))).))))(((.......)))).))). ( -33.70) >DroMoj_CAF1 27494 105 + 1 ACAUAUAUUUGAUUA----GUUAAUCUACCUUAUACCACUCAAUGGUGGGCUCCGGCUUGCCCUCCACUUUGCAAUUGAGCGUGGCGGGUUCAUUCUUCUUGACGACCA ...((((...((((.----...)))).....))))((((....(((.((((........)))).)))....((......))))))..((((...((.....)).)))). ( -22.90) >DroAna_CAF1 6024 92 + 1 ---------------AUGU-AU-CCUUACCUUGUACCACUCGAUGGUGGGCUCCGGCUUGCCCUCCACCUUGCAGUUGAGUGUGGCUGGUUCAUUCUUCUUGACCACCA ---------------....-..-.............((((((((((.((((........)))).)))........)))))))(((.((((.((.......))))))))) ( -25.80) >consensus __________________UGGU_ACUUACCUUAAACCACUCAAUGGUGGGCUCCGGCUUGCCCUCCACUUUGCAAUUGAGCGUGGCGGGUUCAUUCUUCUUGACCACCA ............................((.....((((......)))).....))...((((.(((((((......))).)))).))))................... (-17.00 = -16.95 + -0.05)
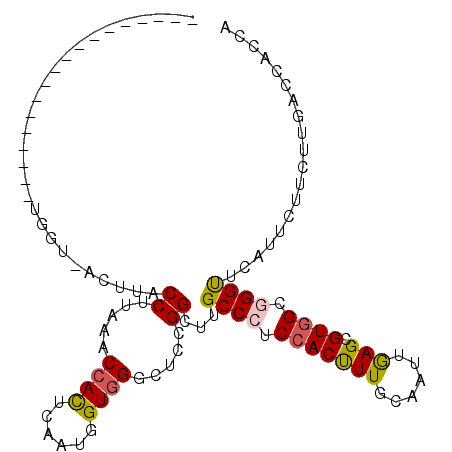
| Location | 18,207,181 – 18,207,274 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.93 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -16.36 |
| Energy contribution | -17.45 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18207181 93 - 20766785 UGGUCGUUAAGAAGAAUGAACCCGCCACGCUCAACUGCAAAGUGGAGGGCAAGCCGGAACCCACCAUUGAGUGGUUUAAGGUACCC-ACCAAAU--------------- ((((((((......))))).(((.((((((......))...)))).)))...(((..((.((((......)))).))..)))....-.)))...--------------- ( -24.90) >DroVir_CAF1 32052 90 - 1 UGGUGGUCAAGAAAAAUGAGCCCGCAACACUCAAUUGCAAGGUGGAGGGCAAGCCGGAGCCCACCAUUGAGUGGUAUAAGGUAUGU-AGC------------------G .((.(.(((.......))).)))((.(((..(.((..(..(((((.((((........)))).)))))..)..)).....)..)))-.))------------------. ( -25.60) >DroWil_CAF1 4872 88 - 1 UGGUGGUUAAGAAGAAUGAACCAGCCACACUUAAUUGUAAAGUGGAAGGCAAACCAGAGCCAACAAUAGAAUGGUUUAAGGUAAGA-A--A------------------ ((((((((..........)))).(((.(((((.......)))))...)))..))))((((((.........)))))).........-.--.------------------ ( -19.10) >DroYak_CAF1 5197 93 - 1 UGGUCGUCAAGAAGAAUGAGCCCGCCACGCUCAACUGCAAAGUGGAGGGCAAGCCGGAACCCACCAUCGAGUGGUUUAAGGUAACC-ACCCAAU--------------- ((((...............((((.((((((......))...)))).))))..(((..((.((((......)))).))..))).)))-)......--------------- ( -28.60) >DroMoj_CAF1 27494 105 - 1 UGGUCGUCAAGAAGAAUGAACCCGCCACGCUCAAUUGCAAAGUGGAGGGCAAGCCGGAGCCCACCAUUGAGUGGUAUAAGGUAGAUUAAC----UAAUCAAAUAUAUGU (((((.(((.......)))(((.(((((((......))..(((((.((((........)))).)))))..)))))....))).)))))..----............... ( -27.10) >DroAna_CAF1 6024 92 - 1 UGGUGGUCAAGAAGAAUGAACCAGCCACACUCAACUGCAAGGUGGAGGGCAAGCCGGAGCCCACCAUCGAGUGGUACAAGGUAAGG-AU-ACAU--------------- (((((((((.......)).)))).)))......((..(..(((((.((((........)))).)))))..)..))...........-..-....--------------- ( -27.10) >consensus UGGUCGUCAAGAAGAAUGAACCCGCCACACUCAACUGCAAAGUGGAGGGCAAGCCGGAGCCCACCAUUGAGUGGUAUAAGGUAAGC_ACCA__________________ ...................(((.(((((((......))..(((((.((((........)))).)))))..)))))....)))........................... (-16.36 = -17.45 + 1.09)

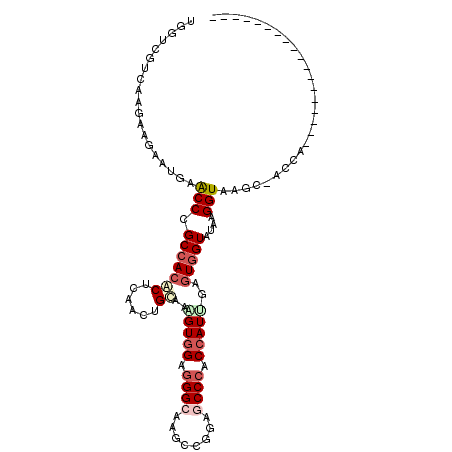
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:27 2006