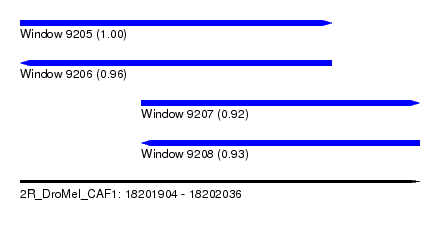
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,201,904 – 18,202,036 |
| Length | 132 |
| Max. P | 0.995182 |

| Location | 18,201,904 – 18,202,007 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.01 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -25.29 |
| Energy contribution | -26.47 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.03 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18201904 103 + 20766785 ACCGAAGAUUCCACUGCAACAUGUUUCCUCUUUAUCUGCUAGCAUCAGUUUUC----------------GAAGACGCAUGUUGUAGUGGACUCCAGACAUCAAUUAUUCUAUUCGAUAA ..((((((.(((((((((((((((.((.((.....(((.......))).....----------------)).)).))))))))))))))).)).(((..........))).)))).... ( -31.10) >DroSec_CAF1 26970 119 + 1 ACCGAAGAUUCCACUGCAACAUGGUUUCUCUUUAUCUGGUAGCAUCCGUUUUCAACUUCUGAAUCUGGAGAACACGCAUGUUGUAGUGGACUCCAGGCAUCAAUUAUUCUAUAUGAUAA .((...((.((((((((((((((((((((((......((......))...((((.....))))...)))))).)).)))))))))))))).))..)).....(((((.....))))).. ( -35.60) >DroSim_CAF1 28575 119 + 1 ACCGAAGAUUCCACUGCAACAUGUUUUCUCUUUAUCUGGUAGCAUCAGUUUUCAACUUCUGAAUCUGGAGAAGACGCAUGUUGUAGUGGACUCCAGGCAUCAAUUAUUCUAUUCGAUAA .((...((.((((((((((((((((((((((....(((((...)))))..((((.....))))...)))))))..))))))))))))))).))..))...................... ( -36.40) >DroEre_CAF1 27644 116 + 1 ACCGAAGAUUCCACUGCAACAUGUUUUCUCUUUAUCUGCUAGCAUCGCUUUUUCAAUUCUCAGUCCAGAGAA---GCAUGUUGUAGUGGGCUCAAGGCAUCAAUUAUUCUAUUCGAUAA ..((((....((((((((((((((((.((((....(((..(((...)))...........)))...))))))---))))))))))))))((.....)).............)))).... ( -34.22) >consensus ACCGAAGAUUCCACUGCAACAUGUUUUCUCUUUAUCUGCUAGCAUCAGUUUUCAACUUCUGAAUCUGGAGAAGACGCAUGUUGUAGUGGACUCCAGGCAUCAAUUAUUCUAUUCGAUAA .((...((.(((((((((((((((.((((((...................................))))))...))))))))))))))).))..))...................... (-25.29 = -26.47 + 1.19)

| Location | 18,201,904 – 18,202,007 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 85.01 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -22.67 |
| Energy contribution | -23.54 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18201904 103 - 20766785 UUAUCGAAUAGAAUAAUUGAUGUCUGGAGUCCACUACAACAUGCGUCUUC----------------GAAAACUGAUGCUAGCAGAUAAAGAGGAAACAUGUUGCAGUGGAAUCUUCGGU .((((((.........)))))).((((((((((((.(((((((..(((((----------------.....(((.......))).....)))))..))))))).))))))..)))))). ( -33.80) >DroSec_CAF1 26970 119 - 1 UUAUCAUAUAGAAUAAUUGAUGCCUGGAGUCCACUACAACAUGCGUGUUCUCCAGAUUCAGAAGUUGAAAACGGAUGCUACCAGAUAAAGAGAAACCAUGUUGCAGUGGAAUCUUCGGU .(((((...........))))).((((((((((((.(((((((.((.(((((....(((((...)))))...((......)).......)))))))))))))).))))))..)))))). ( -33.20) >DroSim_CAF1 28575 119 - 1 UUAUCGAAUAGAAUAAUUGAUGCCUGGAGUCCACUACAACAUGCGUCUUCUCCAGAUUCAGAAGUUGAAAACUGAUGCUACCAGAUAAAGAGAAAACAUGUUGCAGUGGAAUCUUCGGU .((((((.........)))))).((((((((((((.(((((((....(((((....(((((...)))))..(((.......))).....)))))..))))))).))))))..)))))). ( -31.10) >DroEre_CAF1 27644 116 - 1 UUAUCGAAUAGAAUAAUUGAUGCCUUGAGCCCACUACAACAUGC---UUCUCUGGACUGAGAAUUGAAAAAGCGAUGCUAGCAGAUAAAGAGAAAACAUGUUGCAGUGGAAUCUUCGGU ..(((((.........)))))(((..((..(((((.(((((((.---((((((..((((...((((......)))).....))).)..))))))..))))))).)))))..))...))) ( -31.90) >consensus UUAUCGAAUAGAAUAAUUGAUGCCUGGAGUCCACUACAACAUGCGUCUUCUCCAGAUUCAGAAGUUGAAAACUGAUGCUACCAGAUAAAGAGAAAACAUGUUGCAGUGGAAUCUUCGGU .((((((.........)))))).((((((((((((.(((((((....(((((.....................................)))))..))))))).))))))..)))))). (-22.67 = -23.54 + 0.87)


| Location | 18,201,944 – 18,202,036 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -29.21 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.92 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18201944 92 + 20766785 AGCAUCAGUUUUC----------------GAAGACGCAUGUUGUAGUGGACUCCAGACAUCAAUUAUUCUAUUCGAUAACUGAUGCAGAAACUUUGAGUUCACUCAGU .(((((((((.((----------------(((.......((((...(((...))).....)))).......))))).))))))))).......(((((....))))). ( -26.94) >DroSec_CAF1 27010 108 + 1 AGCAUCCGUUUUCAACUUCUGAAUCUGGAGAACACGCAUGUUGUAGUGGACUCCAGGCAUCAAUUAUUCUAUAUGAUAACUGAUGCAGAAACUUUGAGUUCACUCAGU .......(((((............((((((..(((((.....)).)))..))))))((((((.(((((......))))).)))))).))))).(((((....))))). ( -30.70) >DroSim_CAF1 28615 108 + 1 AGCAUCAGUUUUCAACUUCUGAAUCUGGAGAAGACGCAUGUUGUAGUGGACUCCAGGCAUCAAUUAUUCUAUUCGAUAACUGAUGCAGAAACUUUGAGUUCACUCAGU .(((((((((.((..(....)..(((((((....(((........)))..))))))).................)).))))))))).......(((((....))))). ( -30.00) >consensus AGCAUCAGUUUUCAACUUCUGAAUCUGGAGAAGACGCAUGUUGUAGUGGACUCCAGGCAUCAAUUAUUCUAUUCGAUAACUGAUGCAGAAACUUUGAGUUCACUCAGU (((((..((((((................))))))..)))))(.(((((((((.((((((((.(((((......))))).)))))).....))..))))))))))... (-23.26 = -23.92 + 0.67)



| Location | 18,201,944 – 18,202,036 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -22.31 |
| Energy contribution | -22.67 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18201944 92 - 20766785 ACUGAGUGAACUCAAAGUUUCUGCAUCAGUUAUCGAAUAGAAUAAUUGAUGUCUGGAGUCCACUACAACAUGCGUCUUC----------------GAAAACUGAUGCU ..((((....))))........(((((((((.(((((..........(((((.((..((.....))..)).))))))))----------------)).))))))))). ( -28.30) >DroSec_CAF1 27010 108 - 1 ACUGAGUGAACUCAAAGUUUCUGCAUCAGUUAUCAUAUAGAAUAAUUGAUGCCUGGAGUCCACUACAACAUGCGUGUUCUCCAGAUUCAGAAGUUGAAAACGGAUGCU ..((((....)))).(((.((((((((((((((........)))))))))))((((((..(((..........)))..)))))).................))).))) ( -33.00) >DroSim_CAF1 28615 108 - 1 ACUGAGUGAACUCAAAGUUUCUGCAUCAGUUAUCGAAUAGAAUAAUUGAUGCCUGGAGUCCACUACAACAUGCGUCUUCUCCAGAUUCAGAAGUUGAAAACUGAUGCU ..((((....))))........(((((((((.((((...((((....(((((.((..((.....))..)).)))))........)))).....)))).))))))))). ( -30.10) >consensus ACUGAGUGAACUCAAAGUUUCUGCAUCAGUUAUCGAAUAGAAUAAUUGAUGCCUGGAGUCCACUACAACAUGCGUCUUCUCCAGAUUCAGAAGUUGAAAACUGAUGCU ..((((((.((((..((.....((((((((((((.....).))))))))))))).)))).)))).))....(((((.......(((......))).......))))). (-22.31 = -22.67 + 0.36)

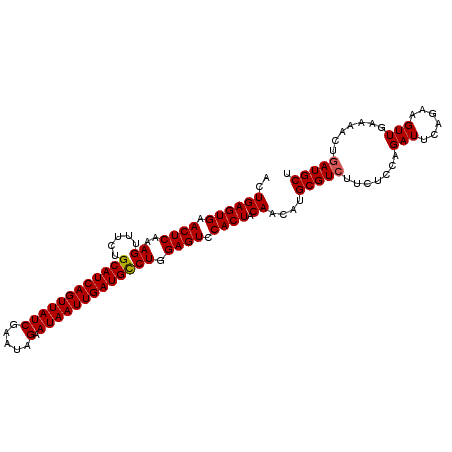
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:24 2006