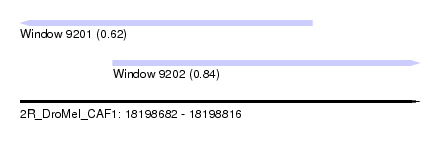
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,198,682 – 18,198,816 |
| Length | 134 |
| Max. P | 0.844998 |

| Location | 18,198,682 – 18,198,780 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.12 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -19.90 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18198682 98 - 20766785 UAUGCACUUCGAAUUCGCUGGGCUGGCCCAAACAUCAUUAUAUGUGCGUGGGCCAUUUCCCUCAUCCUCAGCUGCCUCA---------UCUGCCCACGGAGAUUGCA ..(((((((((.....((((((.(((((((.((((......))))...)))))))...........)))))).((....---------...))...)))))..)))) ( -29.40) >DroSec_CAF1 23795 98 - 1 UAUGCACUUCGAAUUCGCUGGGCUGGCUCAUACAUCGGUAUAUCUGCGUGGGCCAUUUCCCUCAUCCUCAGCUGCCUCA---------UCUGCCCACGGAGAUUGCA ..(((((((((.....((((((.((((((((....(((.....))).))))))))...........)))))).((....---------...))...)))))..)))) ( -25.10) >DroSim_CAF1 25500 98 - 1 UAUGCACUUUGAAUUCGCUGGGCUGGCCCAAACAUCGGUAUAUCUGCGUGGGCCAUUUCCCUCAUCCUCAGCUGCCUCA---------UCUGCCCACGGAGAUUGCA ..((((.......((((.((((((((((((.....(((.....)))..)))))))..............((.((...))---------.)))))))))))...)))) ( -26.60) >DroEre_CAF1 24439 98 - 1 UAUGCACUUCGAAUUCGCUGGGCUGGCCCAAACAUCGCAAUGGCUGCGUGGGCCAUUUCCCUCAUCCUCAGCUGCCUCA---------UCUGCCCACGAAGAUUGCA ..(((((((((.....((((((.(((((((.....((((.....)))))))))))...........)))))).((....---------...))...)))))..)))) ( -30.10) >DroYak_CAF1 25262 107 - 1 UAUGCACUUCAAAUUCGCUGGGCUGACCCAAACAUGGUUAUAUCUGCGUGGGCCAUUUCCCUCAUCCUCAGCUGCCUCAUUUGCCUCGUCUGCCCACGGAGAUUGCA ..((((..((...((((.(((((.(((.((((...(((.....(((.(((((........)))))...)))..)))...))))....))).))))))))))).)))) ( -26.90) >DroPer_CAF1 23306 86 - 1 UAUACAGAUGGAAUCUUCUGGGCUGGCCCAAGCCUCAAGUUUGCUGCGUGGGCCU--CCCCUC---UCUAG-----UCA---------UCUGCCCACAGCCA--GCC ....((((........))))(((((((.(((((.....)))))....((((((..--......---.....-----...---------...)))))).))))--))) ( -30.01) >consensus UAUGCACUUCGAAUUCGCUGGGCUGGCCCAAACAUCGGUAUAUCUGCGUGGGCCAUUUCCCUCAUCCUCAGCUGCCUCA_________UCUGCCCACGGAGAUUGCA ...(((..((...((((.((((((((((((..((..........))..))))))).....((.......))....................))))))))))).))). (-19.90 = -19.98 + 0.09)

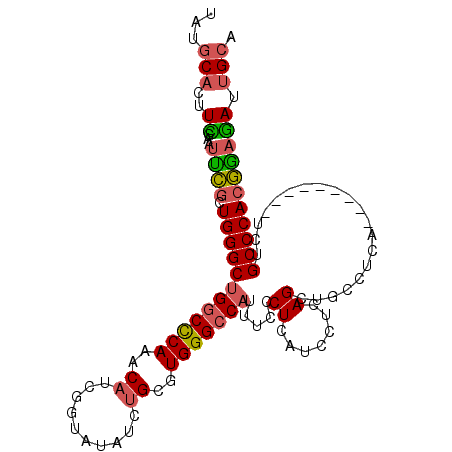
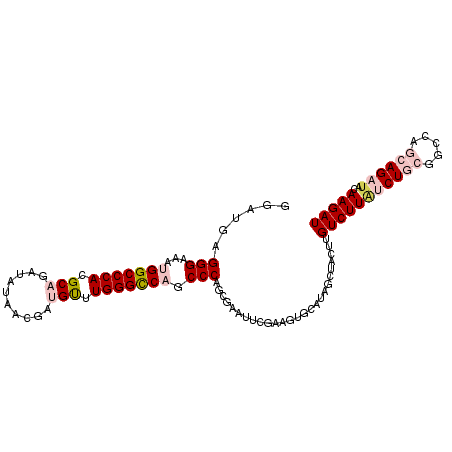
| Location | 18,198,713 – 18,198,816 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -24.95 |
| Energy contribution | -26.32 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.844998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18198713 103 + 20766785 GGAUGAGGGAAAUGGCCCACGCACAUAUAAUGAUGUUUGGGCCAGCCCAGCGAAUUCGAAGUGCAUAGCUACUUGUCUUAUCUGCGGCCAGCAGAUACAAGAU ......(((...(((((((.((((((...))).))).))))))).)))(((................))).(((((...((((((.....))))))))))).. ( -36.69) >DroPse_CAF1 23388 98 + 1 A---GAGGGG--AGGCCCACGCAGCAAGCUUGAGGCUUGGGCCAGCCCAGAAGAUUCCAUCUGUAUAGCUACUCGUCUUGCCUACGGAUAUCAGAUUCAAGAU .---..(((.--.((((((.((..((....))..)).))))))..))).(((......(((((((..((..........)).)))))))......)))..... ( -31.00) >DroSim_CAF1 25531 103 + 1 GGAUGAGGGAAAUGGCCCACGCAGAUAUACCGAUGUUUGGGCCAGCCCAGCGAAUUCAAAGUGCAUAGCUACUUGUCUUAUCUGCGGCCAGCAGAUACAAGAU ......(((...(((((((.(((..........))).))))))).)))(((................))).(((((...((((((.....))))))))))).. ( -35.89) >DroEre_CAF1 24470 103 + 1 GGAUGAGGGAAAUGGCCCACGCAGCCAUUGCGAUGUUUGGGCCAGCCCAGCGAAUUCGAAGUGCAUAGCUACUUGUCUUAUCUGCGGCCAGCAGAUACAAGAU ......(((...(((((((.(((((....))..))).))))))).)))(((................))).(((((...((((((.....))))))))))).. ( -36.59) >DroYak_CAF1 25302 103 + 1 GGAUGAGGGAAAUGGCCCACGCAGAUAUAACCAUGUUUGGGUCAGCCCAGCGAAUUUGAAGUGCAUAGCUACUUGUCUUAUCUGCGGCCAGCAGAUACAAGAU ......(((...(((((((.(((..........))).))))))).)))(((.....((.....))..))).(((((...((((((.....))))))))))).. ( -33.20) >DroPer_CAF1 23330 98 + 1 A---GAGGGG--AGGCCCACGCAGCAAACUUGAGGCUUGGGCCAGCCCAGAAGAUUCCAUCUGUAUAGCUACUCGUCUUGCCUACGGAUAUCAGAAUCAAGAU .---..(((.--.((((((.((..((....))..)).))))))..)))....(((((.(((((((..((..........)).)))))))....)))))..... ( -33.50) >consensus GGAUGAGGGAAAUGGCCCACGCAGAUAUAACGAUGUUUGGGCCAGCCCAGCGAAUUCGAAGUGCAUAGCUACUUGUCUUAUCUGCGGCCAGCAGAUACAAGAU ......(((...(((((((.(((..........))).))))))).)))..........................(((((((((((.....))))))..))))) (-24.95 = -26.32 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:19 2006