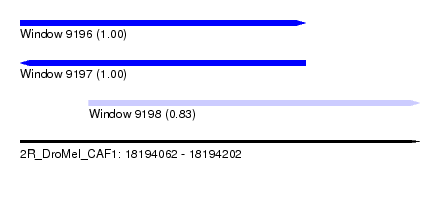
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,194,062 – 18,194,202 |
| Length | 140 |
| Max. P | 0.998915 |

| Location | 18,194,062 – 18,194,162 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -28.94 |
| Consensus MFE | -22.18 |
| Energy contribution | -22.22 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18194062 100 + 20766785 ----------UGCGUUUUACUAGUAUAC------AAAAGUCCCUUUUACAAAAGCUAUGUUUUCUAAGCAUAGAAACUGCAACGAAAC-UUCAGGUCAUGUAAAAGGGACU-UUACCU ----------..................------.(((((((((((((((....(((((((.....)))))))...(((.((......-)))))....)))))))))))))-)).... ( -28.00) >DroSec_CAF1 19353 106 + 1 ----------UGAGUAUUAUUAGUCUACAUUUGCAAAAGUCCCUUUUACAAGAGCUAUGUUUUCUAAGCAUAGAAACUGCAACGAAAC-UUCAGGUCGUGUAAAAGGGGCU-UUACCU ----------.((.((....)).))..........(((((((((((((((.((.((..(((((....(((.......)))...)))))-...)).)).)))))))))))))-)).... ( -30.10) >DroSim_CAF1 19394 106 + 1 ----------UGAGUAUUACUAGUCUACAUUUGCAAAAGUCCCUUUUACAAAAGCUUUGUUUUCUAAGCAUAGAAACUGCAACGAAAC-UUCAGGUCUUGUAAAAGGGACU-UUACCU ----------.........................((((((((((((((((.(.((..(((((....(((.......)))...)))))-...)).).))))))))))))))-)).... ( -27.80) >DroEre_CAF1 19923 118 + 1 AUUAAGCUAUUGCGUUUUACUAGUACACAUUUGCAAGAGUCCCUUUUACAAAAGCUACGUUUUCUAAGCACAGAAACUGCAAUAAAAAUUUCAGGUUGUGUAAAGGGGGCUUUUACCU .....((...((.((.........)).))...))(((((((((((((((....(((..........)))((((...(((.(((....))).))).))))))))))))))))))).... ( -29.80) >DroYak_CAF1 20735 107 + 1 ----------UAAGUUUUACUAGUAUGCAUUUGCAAAAGCCCCUUCUACAAAAGCUAUGUUUUCUCAGCAUGGAAACUGCAAUAAAAC-UUUAGGUGGUGCAAAGGGGGCUUUUACCU ----------................((....))(((((((((((...((..(.(((.(((((....(((.(....))))...)))))-..))).)..))...))))))))))).... ( -29.00) >consensus __________UGAGUUUUACUAGUAUACAUUUGCAAAAGUCCCUUUUACAAAAGCUAUGUUUUCUAAGCAUAGAAACUGCAACGAAAC_UUCAGGUCGUGUAAAAGGGGCU_UUACCU .....................................(((((((((((((....(((((((.....)))))))...(((............)))....)))))))))))))....... (-22.18 = -22.22 + 0.04)

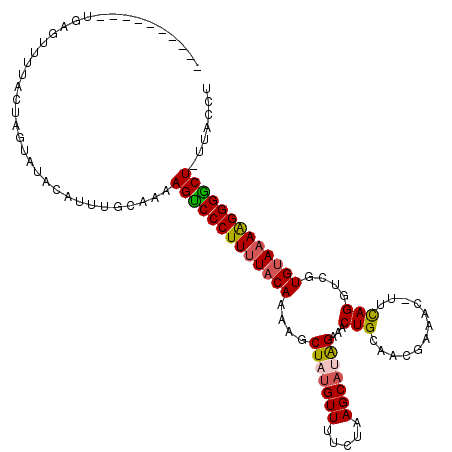
| Location | 18,194,062 – 18,194,162 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -25.62 |
| Energy contribution | -26.14 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18194062 100 - 20766785 AGGUAA-AGUCCCUUUUACAUGACCUGAA-GUUUCGUUGCAGUUUCUAUGCUUAGAAAACAUAGCUUUUGUAAAAGGGACUUUU------GUAUACUAGUAAAACGCA---------- .(((((-(((((((((((((.((.(((..-.((((...(((.......)))...))))...))).)).))))))))))))))..------...))))...........---------- ( -29.70) >DroSec_CAF1 19353 106 - 1 AGGUAA-AGCCCCUUUUACACGACCUGAA-GUUUCGUUGCAGUUUCUAUGCUUAGAAAACAUAGCUCUUGUAAAAGGGACUUUUGCAAAUGUAGACUAAUAAUACUCA---------- ..((((-((.((((((((((.((.(((..-.((((...(((.......)))...))))...))).)).)))))))))).).)))))......................---------- ( -27.60) >DroSim_CAF1 19394 106 - 1 AGGUAA-AGUCCCUUUUACAAGACCUGAA-GUUUCGUUGCAGUUUCUAUGCUUAGAAAACAAAGCUUUUGUAAAAGGGACUUUUGCAAAUGUAGACUAGUAAUACUCA---------- ..((((-((((((((((((((((.((...-.((((...(((.......)))...))))....)).))))))))))))))).))))).......((.((....)).)).---------- ( -34.80) >DroEre_CAF1 19923 118 - 1 AGGUAAAAGCCCCCUUUACACAACCUGAAAUUUUUAUUGCAGUUUCUGUGCUUAGAAAACGUAGCUUUUGUAAAAGGGACUCUUGCAAAUGUGUACUAGUAAAACGCAAUAGCUUAAU ..((((.((.(((.((((((.((.(((...((((((..((((...))))...))))))...))).)).)))))).))).)).))))...(((((.........))))).......... ( -23.80) >DroYak_CAF1 20735 107 - 1 AGGUAAAAGCCCCCUUUGCACCACCUAAA-GUUUUAUUGCAGUUUCCAUGCUGAGAAAACAUAGCUUUUGUAGAAGGGGCUUUUGCAAAUGCAUACUAGUAAAACUUA---------- ..(((((((((((.((((((....(((..-(((((....((((......))))..))))).)))....)))))).)))))))))))...(((......))).......---------- ( -32.60) >consensus AGGUAA_AGCCCCUUUUACACGACCUGAA_GUUUCGUUGCAGUUUCUAUGCUUAGAAAACAUAGCUUUUGUAAAAGGGACUUUUGCAAAUGUAUACUAGUAAAACUCA__________ ..((((.(((((((((((((.((.(((....((((...(((.......)))...))))...))).)).))))))))))))).))))................................ (-25.62 = -26.14 + 0.52)

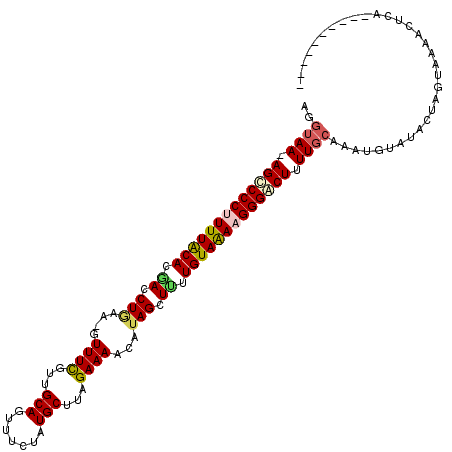
| Location | 18,194,086 – 18,194,202 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -19.22 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18194086 116 + 20766785 CCCUUUUACAAAAGCUAUGUUUUCUAAGCAUAGAAACUGCAACGAAAC-UUCAGGUCAUGUAAAAGGGACU-UUACCUAACUUUGAAGGACCGAAUGUUCUCAAAUAAACUCAAAUAA ((((((((((....(((((((.....)))))))...(((.((......-)))))....))))))))))...-.........(((((.((((.....)))))))))............. ( -25.10) >DroSec_CAF1 19383 108 + 1 CCCUUUUACAAGAGCUAUGUUUUCUAAGCAUAGAAACUGCAACGAAAC-UUCAGGUCGUGUAAAAGGGGCU-UUACCUAACUUUGGAGGACCGAAUGUUCUCAAAUAAA--------A ((((((((((.((.((..(((((....(((.......)))...)))))-...)).)).))))))))))...-.............((((((.....)))))).......--------. ( -28.60) >DroSim_CAF1 19424 108 + 1 CCCUUUUACAAAAGCUUUGUUUUCUAAGCAUAGAAACUGCAACGAAAC-UUCAGGUCUUGUAAAAGGGACU-UUACCUAACUUUGGAGGACCGAAUGUUCUCAAAUAAA--------A (((((((((((.(.((..(((((....(((.......)))...)))))-...)).).)))))))))))...-.............((((((.....)))))).......--------. ( -26.30) >DroEre_CAF1 19963 110 + 1 CCCUUUUACAAAAGCUACGUUUUCUAAGCACAGAAACUGCAAUAAAAAUUUCAGGUUGUGUAAAGGGGGCUUUUACCUAACCUUGAAGGACCGAAUCUUCUCAAAUAAA--------A (((((((((....(((..........)))((((...(((.(((....))).))).)))))))))))))................(((((......))))).........--------. ( -21.10) >DroYak_CAF1 20765 109 + 1 CCCUUCUACAAAAGCUAUGUUUUCUCAGCAUGGAAACUGCAAUAAAAC-UUUAGGUGGUGCAAAGGGGGCUUUUACCUAACCUUGGAGAACUGAGUGUUCACAAAUAAU--------A .............(((..((((((.(((((.(....))))........-.((((((((.((.......))..))))))))...))))))))..))).............--------. ( -22.00) >consensus CCCUUUUACAAAAGCUAUGUUUUCUAAGCAUAGAAACUGCAACGAAAC_UUCAGGUCGUGUAAAAGGGGCU_UUACCUAACUUUGGAGGACCGAAUGUUCUCAAAUAAA________A ((((((((((....(((((((.....)))))))...(((............)))....))))))))))..............(((.(((((.....)))))))).............. (-19.22 = -19.90 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:15 2006