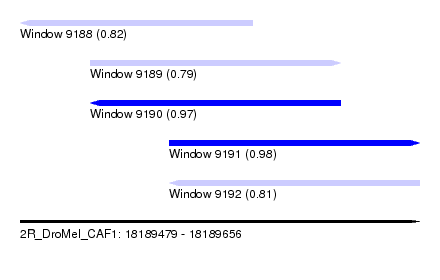
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,189,479 – 18,189,656 |
| Length | 177 |
| Max. P | 0.975028 |

| Location | 18,189,479 – 18,189,582 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 89.71 |
| Mean single sequence MFE | -23.21 |
| Consensus MFE | -17.25 |
| Energy contribution | -18.13 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18189479 103 - 20766785 CAGCUAGAGCU-GUAGCUGUAGCUGUAGCACCACCGAAACACAAAGCAGCACCACCAUCUGCAAUCGCAGCAAGGUUAUCACAGCAGCCACACACACGCACAC------A-- ((((....)))-)..(((((.(((((...................)))))...(((..((((....))))...)))....)))))..................------.-- ( -23.81) >DroSec_CAF1 14728 101 - 1 CAGCUAGAGCU-GUAGCUGUAGCUGUAGCACCACCGAAACACAAAGCAGCACCACCAUCUGCAAUCGCAGCAAGGUUAUCACAGCAGCCACACACA--CACAC------A-- ((((....)))-)..(((((.(((((...................)))))...(((..((((....))))...)))....)))))...........--.....------.-- ( -23.81) >DroSim_CAF1 14775 93 - 1 CAGCUAGAGCU-GUA------GCUGUAGCACCACCGAAACACAAAGCAGCACCACCAUCUGCAAUCGCAGCAAGGUUAUCACAGCAGCCACACAC----ACAC------A-- ..(((...(((-((.------(((((...................)))))...(((..((((....))))...)))....)))))))).......----....------.-- ( -20.41) >DroEre_CAF1 15184 99 - 1 CAGCUAGAGCU-GUAGCUGUAGCUGUAGCACCACCGAAACACAAAGCAGCACCACCAUCUGCAAUCGUGGCAAGGUUAUCGCAGCAGCCACACACA----UAC------A-- ((((((.(((.-...))).))))))....................((((.........))))....(((((...(((.....))).))))).....----...------.-- ( -26.40) >DroYak_CAF1 15869 108 - 1 CAGCUAGAACUCGUAGCUGUAGCUGUAGCACCACCAAAACACAAAGCAGCACCACCAUCUGCAAUCGCAGCAAGGUUAUCACAGCAGCCACACACA----CACGCAUACACA ..((..(....)((.(((((.(((((...................)))))...(((..((((....))))...))).......))))).)).....----...))....... ( -21.61) >consensus CAGCUAGAGCU_GUAGCUGUAGCUGUAGCACCACCGAAACACAAAGCAGCACCACCAUCUGCAAUCGCAGCAAGGUUAUCACAGCAGCCACACACA___ACAC______A__ .(((....))).((.(((((.(((((...................)))))...(((..((((....))))...))).......))))).))..................... (-17.25 = -18.13 + 0.88)

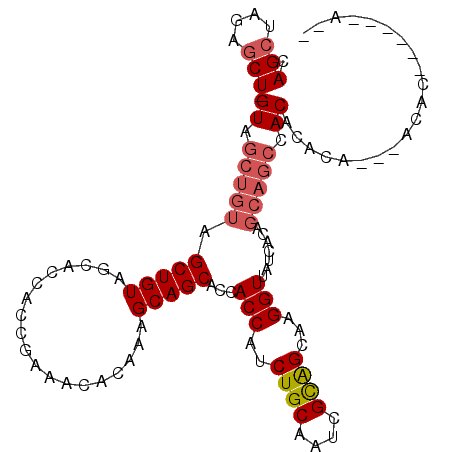
| Location | 18,189,510 – 18,189,621 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -38.72 |
| Consensus MFE | -32.28 |
| Energy contribution | -33.52 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18189510 111 + 20766785 UUGCUGCGAUUGCAGAUGGUGGUGCUGCUUUGUGUUUCGGUGGUGCUACAGCUACAGCUAC-AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCCAUGGUCAC .(((.......)))(((.((((........((((...(((.((..(((((((((((((((.-.....)))))))))))....))))..))))))))).....)))).))).. ( -40.92) >DroSec_CAF1 14757 111 + 1 UUGCUGCGAUUGCAGAUGGUGGUGCUGCUUUGUGUUUCGGUGGUGCUACAGCUACAGCUAC-AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCCUUGGUCAC ....(((((..((.....((((..(..((.........))..)..)))).(((((((((((-(((....)))))))))....))))))).)))))...((......)).... ( -39.40) >DroSim_CAF1 14802 105 + 1 UUGCUGCGAUUGCAGAUGGUGGUGCUGCUUUGUGUUUCGGUGGUGCUACAGC------UAC-AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCCUUGGUCAC ....(((((..(((....((((..(..((.........))..)..))))(((------(((-(((....)))))))))........))).)))))...((......)).... ( -36.60) >DroEre_CAF1 15211 111 + 1 UUGCCACGAUUGCAGAUGGUGGUGCUGCUUUGUGUUUCGGUGGUGCUACAGCUACAGCUAC-AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCGUUGGUCAC ..(((((((......((((.((..(..((..........(((((......)))))((((((-(((....)))))))))...))..)..)).).))).....))).))))... ( -41.00) >DroYak_CAF1 15904 112 + 1 UUGCUGCGAUUGCAGAUGGUGGUGCUGCUUUGUGUUUUGGUGGUGCUACAGCUACAGCUACGAGUUCUAGCUGCGGCUAAUGGUGGUGCCUCGCAUAUCCUUUCUUGGUCAC ....(((((..(((....((((..(..((.........))..)..))))(((..((((((.(....)))))))..)))........))).)))))...((......)).... ( -35.70) >consensus UUGCUGCGAUUGCAGAUGGUGGUGCUGCUUUGUGUUUCGGUGGUGCUACAGCUACAGCUAC_AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCCUUGGUCAC .(((..(((..(((.(.(((......))).).))).)))(.((..(((((((((((((((.......)))))))))))....))))..)).))))...((......)).... (-32.28 = -33.52 + 1.24)

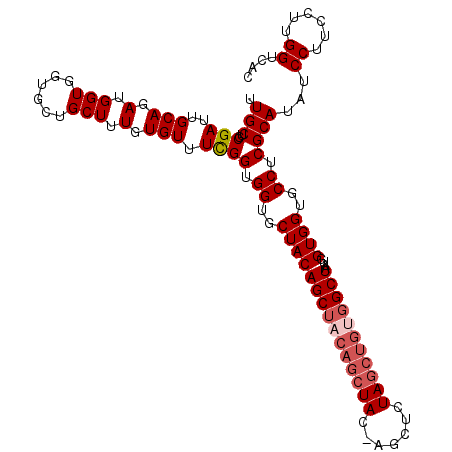
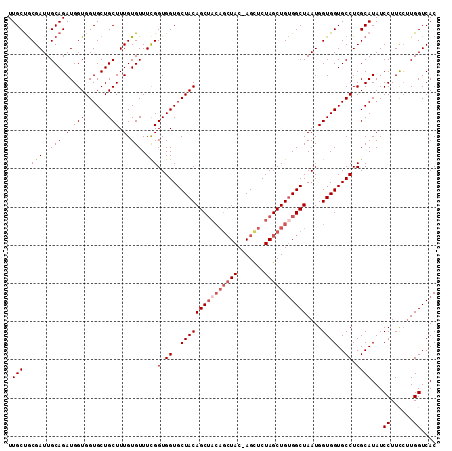
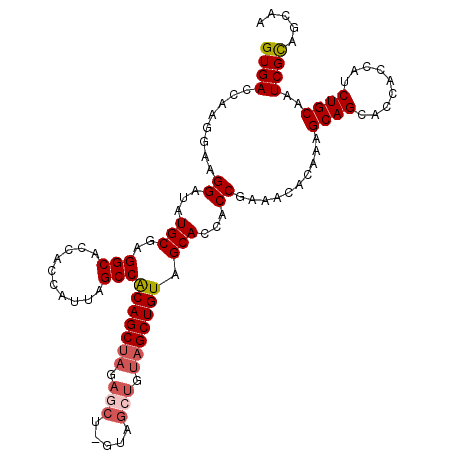
| Location | 18,189,510 – 18,189,621 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -30.86 |
| Consensus MFE | -26.38 |
| Energy contribution | -27.26 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18189510 111 - 20766785 GUGACCAUGGAAGGAUAUGCGAGGCACCACCAUUAGCCACAGCUAGAGCU-GUAGCUGUAGCUGUAGCACCACCGAAACACAAAGCAGCACCACCAUCUGCAAUCGCAGCAA ......((((..((...(((..((.....))....((.(((((((.(((.-...))).))))))).))................)))...)).))))((((....))))... ( -32.70) >DroSec_CAF1 14757 111 - 1 GUGACCAAGGAAGGAUAUGCGAGGCACCACCAUUAGCCACAGCUAGAGCU-GUAGCUGUAGCUGUAGCACCACCGAAACACAAAGCAGCACCACCAUCUGCAAUCGCAGCAA ((((....(((.((...(((..((.....))....((.(((((((.(((.-...))).))))))).))...................)))...)).)))....))))..... ( -32.30) >DroSim_CAF1 14802 105 - 1 GUGACCAAGGAAGGAUAUGCGAGGCACCACCAUUAGCCACAGCUAGAGCU-GUA------GCUGUAGCACCACCGAAACACAAAGCAGCACCACCAUCUGCAAUCGCAGCAA ((((....(((.((...(((..(((..........)))...((((.(((.-...------))).))))...................)))...)).)))....))))..... ( -26.50) >DroEre_CAF1 15211 111 - 1 GUGACCAACGAAGGAUAUGCGAGGCACCACCAUUAGCCACAGCUAGAGCU-GUAGCUGUAGCUGUAGCACCACCGAAACACAAAGCAGCACCACCAUCUGCAAUCGUGGCAA .((.(((.(((.((...(((..(((..........)))(((((((.(((.-...))).))))))).)))...))..........((((.........))))..)))))))). ( -35.00) >DroYak_CAF1 15904 112 - 1 GUGACCAAGAAAGGAUAUGCGAGGCACCACCAUUAGCCGCAGCUAGAACUCGUAGCUGUAGCUGUAGCACCACCAAAACACAAAGCAGCACCACCAUCUGCAAUCGCAGCAA ((((........((...(((..((.....))..((((..(((((((....).))))))..))))..)))...))..........((((.........))))..))))..... ( -27.80) >consensus GUGACCAAGGAAGGAUAUGCGAGGCACCACCAUUAGCCACAGCUAGAGCU_GUAGCUGUAGCUGUAGCACCACCGAAACACAAAGCAGCACCACCAUCUGCAAUCGCAGCAA ((((........((...(((..(((..........)))(((((((.(((.....))).))))))).)))...))..........((((.........))))..))))..... (-26.38 = -27.26 + 0.88)

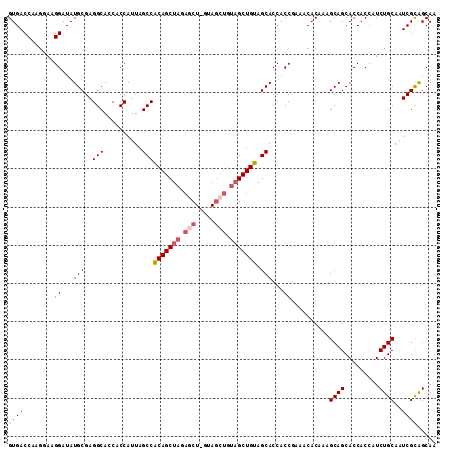
| Location | 18,189,545 – 18,189,656 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -37.92 |
| Consensus MFE | -34.38 |
| Energy contribution | -35.46 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18189545 111 + 20766785 UUCGGUGGUGCUACAGCUACAGCUAC-AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCCAUGGUCACCUUGGGCGGCAUUCACGAAAUGUGUAUCCAUGACA ..(((.((..(((((((((((((((.-.....)))))))))))....))))..)))))..........(((((.(((.((...((.......)).)).)))...)))))... ( -39.20) >DroSec_CAF1 14792 111 + 1 UUCGGUGGUGCUACAGCUACAGCUAC-AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCCUUGGUCACCUUGGGCGGCAUUCACGAAAUGUGUAUCCAUGACA ....((((.((((((((((.(((...-.))).)))))))))).....((((((((.(.((...((......))......)).).)))).))))...........)))).... ( -38.70) >DroSim_CAF1 14837 105 + 1 UUCGGUGGUGCUACAGC------UAC-AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCCUUGGUCACCUUGGGCGGCAUUCACGAAAUGUGUAUCCAUGACA ....(((((((.(((((------(((-(((....)))))))))....((((((((.(.((...((......))......)).).)))).)))).....))))).)))).... ( -36.30) >DroEre_CAF1 15246 111 + 1 UUCGGUGGUGCUACAGCUACAGCUAC-AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCGUUGGUCACCUUGGGCGGCAUUCACGAAAUGUGUAUCCAUGACA ...((.((..(((((((((((((((.-.....)))))))))))....))))..))..((((((..(((((..(((.((...)).)))....)))))))))))..))...... ( -40.70) >DroYak_CAF1 15939 112 + 1 UUUGGUGGUGCUACAGCUACAGCUACGAGUUCUAGCUGCGGCUAAUGGUGGUGCCUCGCAUAUCCUUUCUUGGUCACCUUGGGCGGCAUUCACGAAAUGUGUAUCCAUGACA ....(((((((.(((((..((((((.(....)))))))..)))....((((((((.(.((...((......))......)).).)))).)))).....))))).)))).... ( -34.70) >consensus UUCGGUGGUGCUACAGCUACAGCUAC_AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCCUUGGUCACCUUGGGCGGCAUUCACGAAAUGUGUAUCCAUGACA ....((((.((((((((((.(((.....))).)))))))))).....((((((((.(.((...((......))......)).).)))).))))...........)))).... (-34.38 = -35.46 + 1.08)

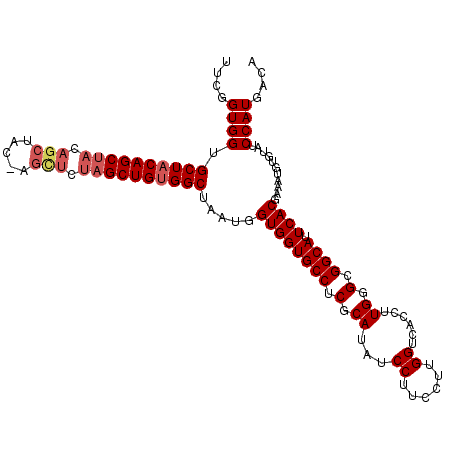
| Location | 18,189,545 – 18,189,656 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -30.88 |
| Energy contribution | -31.92 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18189545 111 - 20766785 UGUCAUGGAUACACAUUUCGUGAAUGCCGCCCAAGGUGACCAUGGAAGGAUAUGCGAGGCACCACCAUUAGCCACAGCUAGAGCU-GUAGCUGUAGCUGUAGCACCACCGAA ..((((((...(((.....)))...(((......)))..))))))..((...(((..(((..........)))(((((((.(((.-...))).))))))).)))...))... ( -37.00) >DroSec_CAF1 14792 111 - 1 UGUCAUGGAUACACAUUUCGUGAAUGCCGCCCAAGGUGACCAAGGAAGGAUAUGCGAGGCACCACCAUUAGCCACAGCUAGAGCU-GUAGCUGUAGCUGUAGCACCACCGAA ..(((((((.......)))))))...........((((.((......))...(((..(((..........)))(((((((.(((.-...))).))))))).))).))))... ( -35.50) >DroSim_CAF1 14837 105 - 1 UGUCAUGGAUACACAUUUCGUGAAUGCCGCCCAAGGUGACCAAGGAAGGAUAUGCGAGGCACCACCAUUAGCCACAGCUAGAGCU-GUA------GCUGUAGCACCACCGAA ..(((((((.......)))))))...........((((.((......))...(((..((.....))..((((.(((((....)))-)).------))))..))).))))... ( -28.50) >DroEre_CAF1 15246 111 - 1 UGUCAUGGAUACACAUUUCGUGAAUGCCGCCCAAGGUGACCAACGAAGGAUAUGCGAGGCACCACCAUUAGCCACAGCUAGAGCU-GUAGCUGUAGCUGUAGCACCACCGAA ..(((((((.......)))))))...........((((.((......))...(((..(((..........)))(((((((.(((.-...))).))))))).))).))))... ( -35.50) >DroYak_CAF1 15939 112 - 1 UGUCAUGGAUACACAUUUCGUGAAUGCCGCCCAAGGUGACCAAGAAAGGAUAUGCGAGGCACCACCAUUAGCCGCAGCUAGAACUCGUAGCUGUAGCUGUAGCACCACCAAA ..(((((((.......)))))))...........((((.((......))...(((..((.....))..((((..(((((((....).))))))..))))..))).))))... ( -31.40) >consensus UGUCAUGGAUACACAUUUCGUGAAUGCCGCCCAAGGUGACCAAGGAAGGAUAUGCGAGGCACCACCAUUAGCCACAGCUAGAGCU_GUAGCUGUAGCUGUAGCACCACCGAA ..(((((((.......)))))))...........((((.((......))...(((..(((..........)))(((((((.(((.....))).))))))).))).))))... (-30.88 = -31.92 + 1.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:09 2006