
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,163,356 – 18,163,466 |
| Length | 110 |
| Max. P | 0.930589 |

| Location | 18,163,356 – 18,163,466 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.12 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -25.38 |
| Energy contribution | -27.30 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18163356 110 - 20766785 GAAACCGUGGAGAUGACAGGAGCACUGGGCCAGCUCAGUUGCCUCUUGUCCACUUUAUCCUGGUAGGAAACUUCGGAAUUCGGAAAGGAAUCUAAGUCCAUGGAUUAACA ....(((((((...(((((((((((((((....))))).)).)))))))).......((((....(((..(....)..)))....)))).......)))))))....... ( -38.30) >DroGri_CAF1 7639 98 - 1 GAAACGGUGCAAAUGACCGGCGCUUUGGGCCAGCUCAGUUGUCUGCUCUCCGUUUUGUCCUGGUAAGCAA---UGGAAGUUUAAA---------CAGCCAACAGUUAUAU (((((((.(((.(..((.(((((.....))..)).).))..).)))...)))))))...(((.(((((..---.....)))))..---------)))............. ( -21.00) >DroSec_CAF1 10930 110 - 1 GAAACCGUGGAGAUGACAGGAGCACUGGGCCAGCUCAGCUGCCUCUUGUCCACUUUGUCCUGGUAGGAAACUCAGGAAUUCGGAAAGGUGUCUAAGUCCAUGGAUUAACU ....(((((((...(((((((((((((((....))))).)).))))))))((((((.(((((..((....))))))).......))))))......)))))))....... ( -43.60) >DroSim_CAF1 10824 99 - 1 GAAACCGUGGAGAUGACAGGAGCACUGGGCCAGCUCAGCUGCCUCUUGUCCACUUUGUCCUGGUAGGAAACUCGGGAAUUUGGAAAGG-----------AUGGGUUAACU ..(((((((((.....(((((((((((((....))))).)).)))))))))))....(((((..((....)))))))...........-----------...)))).... ( -33.30) >DroEre_CAF1 7446 110 - 1 GAAACCGUGGAGAUGACAGGAGCACUGGGCCAGCUCAGCUGCCUCCUGUCCGCCUUGUCCUGGUAGGAAGCUCUAGAAUUCUGAAUGGAAUCUAUAUCCAUGGACUAACG ....(((((((...(((((((((((((((....))))).)).)))))))).((....(((.....))).))..((((.((((....))))))))..)))))))....... ( -40.80) >DroYak_CAF1 6166 106 - 1 GAAACCGUGGAGAUGACAGGAGCGCUGGGCCAGCUCAGCUGCCUCUUGUCCACGUUGUCCUGGUAGGAAACUCCUGAAUUCUAAGCGG----UUAGUCCAUGGAUUAACU ....(((((((...(((((((((((((((....)))))).).))))))))..((((.((..((.((....)))).))......)))).----....)))))))....... ( -40.60) >consensus GAAACCGUGGAGAUGACAGGAGCACUGGGCCAGCUCAGCUGCCUCUUGUCCACUUUGUCCUGGUAGGAAACUCCGGAAUUCGGAAAGG____UAAGUCCAUGGAUUAACU ....(((((((...(((((((((((((((....))))).)).)))))))).........((((.((....))))))....................)))))))....... (-25.38 = -27.30 + 1.92)

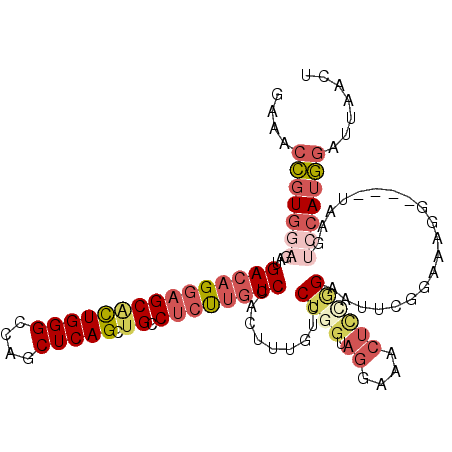
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:58 2006