
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,162,738 – 18,162,855 |
| Length | 117 |
| Max. P | 0.517149 |

| Location | 18,162,738 – 18,162,855 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -40.40 |
| Consensus MFE | -27.54 |
| Energy contribution | -26.40 |
| Covariance contribution | -1.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
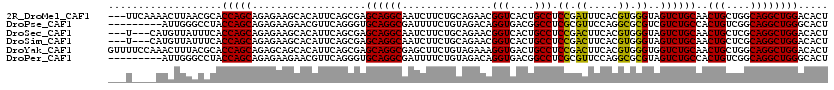
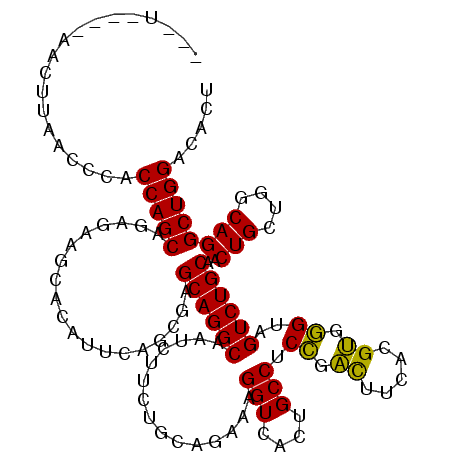
>2R_DroMel_CAF1 18162738 117 + 20766785 ---UUCAAAACUUAACGCACCAGCAGAGAAGCACAUUCAGCGAGCAGGCAAUCUUCUGCAGAACGGUCACUGCCUCCGAUUUCACGUGGGUAGUCUGCAACUGCUGGCAGGCUGGACACU ---................(((((...(((.....))).((.((((((((......)))....(((..((((((..((......))..)))))))))...))))).))..)))))..... ( -37.70) >DroPse_CAF1 5471 111 + 1 ---------AUUGGGCCUACCAGCAGAGAAGAACGUUCAGGGUGCAGGCGAUUUUCUGUAGACAGGUGACGGCCUCGCGUUCCAGGCGCGUCGUCUGCCACUGUCGGCAGGCUGGGCACU ---------.((.(((((.((.((((((((...((((.........)))).)))))))).(((((..((((((...((((.....)))))))))).....))))))).))))).)).... ( -40.90) >DroSec_CAF1 10317 114 + 1 ---U---CAUGUUAUUUCACCAGCAGAGAAGCACAUUCAGCGAGCAGGCAAUCUUCUGCAGAACGGUCACUGCCUCCGACUUCACGUGGGUAGUCUGCAACUGCUCGCAGGCUGGACACU ---.---............(((((...(((.....))).(((((((((((......)))....(((..((((((..((......))..)))))))))...))))))))..)))))..... ( -41.50) >DroSim_CAF1 10211 114 + 1 ---U---CAUGUUAUUUCACCAGCAGAGAAGCACAUUCAGCGAGCAGGCAAUCUUCUGCAGAACGGUCACUGCCUCCGACUUCACGUGGGUAGUCUGCAACUGCUCGCAGGCUGGACACU ---.---............(((((...(((.....))).(((((((((((......)))....(((..((((((..((......))..)))))))))...))))))))..)))))..... ( -41.50) >DroYak_CAF1 5547 120 + 1 GUUUUCCAAACUUUACGCACCAGCAGAGCAGCACAUUCAGCGAGCAGGCGAGCUUCUGUAGAAAGGUGACUGCCUCCGACUUCACGUGGGUGGUCUGCAACUGCUGGCAGGCUGGACACU ....((((........((.(((((((.(((((((...(((.((((......))))))).......)))((..((..((......))..))..))))))..)))))))...)))))).... ( -38.00) >DroPer_CAF1 8647 111 + 1 ---------AUUGGGCCUACCAGCAGAGAAGAACGUUCAGGGUGCAGGCGAUUUUCUGUAGACAGGUGACGGCCUCGCGUUCCAGGCGCGUAGUCUGCCACUGUCGGCAGGCUGGGCACU ---------.....((((.........(((.....))).(..(((((((..((.((((....)))).))..)))).)))..).))))((.(((((((((......))))))))).))... ( -42.80) >consensus ___U____AACUUAACCCACCAGCAGAGAAGCACAUUCAGCGAGCAGGCAAUCUUCUGCAGAAAGGUCACUGCCUCCGACUUCACGUGGGUAGUCUGCAACUGCUGGCAGGCUGGACACU ...................(((((...................((((((...............(((....))).((.((.....)).))..))))))..(((....))))))))..... (-27.54 = -26.40 + -1.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:57 2006