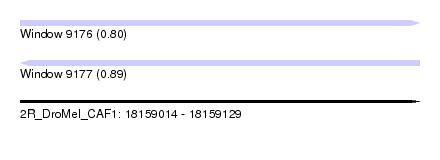
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,159,014 – 18,159,129 |
| Length | 115 |
| Max. P | 0.890353 |

| Location | 18,159,014 – 18,159,129 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.35 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -22.38 |
| Energy contribution | -22.94 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18159014 115 + 20766785 UUCUCUCUGCAUACCGAUGCAACUUGACUUACUUGACAAGCAAGUUAAGCAUGUGGUAUCCAUCUGCUUGCCAUUGGGCAAAGGGUAAAAAUCCAU-----UGUUAUUUUGCAGAUCGAU .((..((((((..(((((((((((((.(((.......)))))))))(((((.((((...)))).)))))).))))))((((.((((....)))).)-----))).....))))))..)). ( -35.90) >DroSec_CAF1 6727 115 + 1 UACUCUCUGCAUACCGAUGCAACUUGACUUACUUGACAAGCAAGUAAAGCAUGUGGUAUACAGCUGAUUGCCAUUGGGCAAAGGGUAAAAAUCUAU-----UGUUAUUUUGCAGAUCUGU .....(((((((((((((((.(((((.(((.......))))))))...)))).))))))...(((..(((((....)))))..)))..........-----.........)))))..... ( -30.60) >DroSim_CAF1 6454 111 + 1 UACUCUCUGCAUACCGAUGCAACU---------UGACAAGCAAGUAAAGCAUGUGGUAUACAGCAGAUUGCCAUUGGGCCAAGGGUAAAAAUUUAUAAAUCUAUUAUUUUGCAGAUCUGU ......(((.((((((((((.(((---------((.....)))))...)))).)))))).)))(((((((((....))).....((((((....(((....)))..)))))).)))))). ( -27.10) >consensus UACUCUCUGCAUACCGAUGCAACUUGACUUACUUGACAAGCAAGUAAAGCAUGUGGUAUACAGCUGAUUGCCAUUGGGCAAAGGGUAAAAAUCUAU_____UGUUAUUUUGCAGAUCUGU .....(((((((((((((((.(((((..............)))))...)))).))))))........(((((....))))).((((....))))................)))))..... (-22.38 = -22.94 + 0.56)

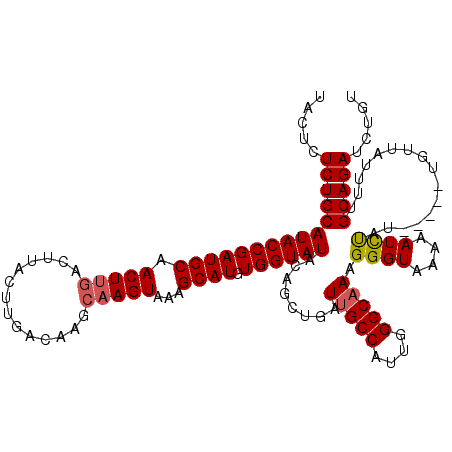
| Location | 18,159,014 – 18,159,129 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.35 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -21.47 |
| Energy contribution | -23.80 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18159014 115 - 20766785 AUCGAUCUGCAAAAUAACA-----AUGGAUUUUUACCCUUUGCCCAAUGGCAAGCAGAUGGAUACCACAUGCUUAACUUGCUUGUCAAGUAAGUCAAGUUGCAUCGGUAUGCAGAGAGAA .((.((((((.........-----..((........)).(((((....)))))))))))(.(((((..((((..((((((((((.....)))).)))))))))).))))).).))..... ( -32.60) >DroSec_CAF1 6727 115 - 1 ACAGAUCUGCAAAAUAACA-----AUAGAUUUUUACCCUUUGCCCAAUGGCAAUCAGCUGUAUACCACAUGCUUUACUUGCUUGUCAAGUAAGUCAAGUUGCAUCGGUAUGCAGAGAGUA ..(((((((..........-----.))))))).(((.(((((((....)))))....(((((((((..((((...(((((((((.....)))).))))).)))).))))))))))).))) ( -31.70) >DroSim_CAF1 6454 111 - 1 ACAGAUCUGCAAAAUAAUAGAUUUAUAAAUUUUUACCCUUGGCCCAAUGGCAAUCUGCUGUAUACCACAUGCUUUACUUGCUUGUCA---------AGUUGCAUCGGUAUGCAGAGAGUA ..(((((((........))))))).................(((....)))..(((.(((((((((..((((...(((((.....))---------))).)))).))))))))))))... ( -30.20) >consensus ACAGAUCUGCAAAAUAACA_____AUAGAUUUUUACCCUUUGCCCAAUGGCAAUCAGCUGUAUACCACAUGCUUUACUUGCUUGUCAAGUAAGUCAAGUUGCAUCGGUAUGCAGAGAGUA .....(((((.............................(((((....)))))........(((((..((((...(((((((((.....)))).))))).)))).))))))))))..... (-21.47 = -23.80 + 2.33)

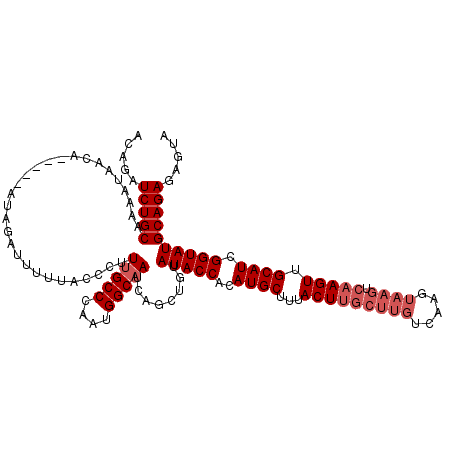
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:55 2006