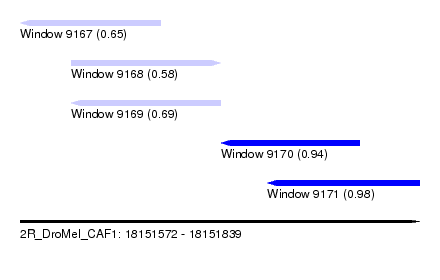
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,151,572 – 18,151,839 |
| Length | 267 |
| Max. P | 0.977631 |

| Location | 18,151,572 – 18,151,666 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.46 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -12.06 |
| Energy contribution | -10.90 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18151572 94 - 20766785 ---GGAUCUAUAUGA---------AACG--CAUG--UGUAUUGGGGUCUGGUGGCAA---UCUCAUGUGCCACACAUGU-----UC--CAGUGACCACAGAAAUUCUGGACGUACUUCUA ---((...((((((.---------....--))))--))((((((..(.((((((((.---.......)))))).)).).-----.)--))))).)).(((.....)))............ ( -26.70) >DroPse_CAF1 5745 118 - 1 UAGGAAUCUACAUAUGUGUGCCUGCACGUACACA--UGCGUCGGUGUCUGGUGGCAGUCGUCUCAUGUGCCACAUAUGUAUGUUUCCUCUAUAGCCACAUAAAUUCUGGACGCACUUCUA .(((((.(((((((((((((....)))...((((--((.(.(((((((....)))).))).).))))))..))))))))).).))))).....((((.........))).)......... ( -33.80) >DroSim_CAF1 49 96 - 1 ---GGAUCUGUAUGG---------UAGG--CAUGUGUGUAUUGGGGUCUGGUGGCAA---UCUCAUGUGCCACACAUGU-----UC--CAGUGACCACAGAAAUUCUGGACGUACUUCUA ---(((...(((((.---------((((--..((((..((((((..(.((((((((.---.......)))))).)).).-----.)--)))))..)))).....))))..))))).))). ( -32.90) >DroYak_CAF1 49 94 - 1 ---GGAUCUAUAUGG---------AAGG--CAUG--UGUAUUGGGGUCUGGUGGCAA---UCUCAUGUGCCACACAUGU-----UC--CAGUGACCACAGAAUUUCUGGACGUACUUCUA ---.........(((---------(((.--.(((--(.((((((..(.((((((((.---.......)))))).)).).-----.)--)))))....(((.....))).)))).)))))) ( -30.10) >DroPer_CAF1 5832 118 - 1 UAGGAAUCUACAUAUGUGUUCCUGCACGUACACA--UGUGUUGGUGUCUGGUGGCAGUCGUCUCAUGUGCCACAUAUGUAUGUUUCCUCUAUAGCCACAUAAAUUCUAGACGCACUUCUA ..((((.(.((((((((((.(......).)))))--))))).)(((((((((((((...........)))))).(((((..(((........))).))))).....)))))))..)))). ( -32.70) >consensus ___GGAUCUAUAUGG_________AACG__CAUG__UGUAUUGGGGUCUGGUGGCAA___UCUCAUGUGCCACACAUGU_____UC__CAGUGACCACAGAAAUUCUGGACGUACUUCUA .....................................((((....((((.((((((...........)))))).(((.............)))..............))))))))..... (-12.06 = -10.90 + -1.16)

| Location | 18,151,606 – 18,151,706 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.49 |
| Mean single sequence MFE | -27.31 |
| Consensus MFE | -11.33 |
| Energy contribution | -11.25 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18151606 100 + 20766785 -ACAUGUGUGGCACAUGAGA---UUGCCACCAGACCCCAAUACA--CAUG--CGUU---------UCAUAUAGAUCC---UAUUGCUUUCAUUAAUGUAUACACAGCGAUUGUGGGGUCU -...((.((((((.......---.))))))))(((((((.....--....--.(..---------((.....))..)---.((((((.................))))))..))))))). ( -25.93) >DroPse_CAF1 5785 117 + 1 UACAUAUGUGGCACAUGAGACGACUGCCACCAGACACCGACGCA--UGUGUACGUGCAGGCACACAUAUGUAGAUUCCUAUAUUGCUUUCAUUAAUAUAUACAAAGUGAAUAUGGGAAC- ((((((((((((...........(((....)))........(((--((....)))))..)).))))))))))..(((((((((((((((.............))))).)))))))))).- ( -34.62) >DroSim_CAF1 83 102 + 1 -ACAUGUGUGGCACAUGAGA---UUGCCACCAGACCCCAAUACACACAUG--CCUA---------CCAUACAGAUCC---UAUUGCUUUCAUUAAUGUAUACACAGCGAUUGUGGGGUCU -...((.((((((.......---.))))))))(((((((........(((--....---------.)))........---.((((((.................))))))..))))))). ( -24.73) >DroYak_CAF1 83 100 + 1 -ACAUGUGUGGCACAUGAGA---UUGCCACCAGACCCCAAUACA--CAUG--CCUU---------CCAUAUAGAUCC---UAUUGCUUUCAUUAAUGUAUACACAGCGAUUGUGGGGCCU -...((.((((((.......---.))))))))(.(((((.....--.(((--....---------.)))........---.((((((.................))))))..))))).). ( -19.73) >DroPer_CAF1 5872 117 + 1 UACAUAUGUGGCACAUGAGACGACUGCCACCAGACACCAACACA--UGUGUACGUGCAGGAACACAUAUGUAGAUUCCUAUAUUGCUUUCAUUAAUAUAUACAAAGUGAAUAUGGGAAC- ((((((((((.(....)......((((.((...((((.......--.))))..))))))...))))))))))..(((((((((((((((.............))))).)))))))))).- ( -31.52) >consensus _ACAUGUGUGGCACAUGAGA___UUGCCACCAGACCCCAAUACA__CAUG__CGUG_________ACAUAUAGAUCC___UAUUGCUUUCAUUAAUGUAUACACAGCGAUUGUGGGGACU .......((((((...........))))))....(((((((......)).................................(((((.................)))))...)))))... (-11.33 = -11.25 + -0.08)

| Location | 18,151,606 – 18,151,706 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.49 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -13.97 |
| Energy contribution | -14.21 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18151606 100 - 20766785 AGACCCCACAAUCGCUGUGUAUACAUUAAUGAAAGCAAUA---GGAUCUAUAUGA---------AACG--CAUG--UGUAUUGGGGUCUGGUGGCAA---UCUCAUGUGCCACACAUGU- ((((((((....((((((((...(((....((........---...))...))).---------.)))--)).)--))...)))))))).((((((.---.......))))))......- ( -30.40) >DroPse_CAF1 5785 117 - 1 -GUUCCCAUAUUCACUUUGUAUAUAUUAAUGAAAGCAAUAUAGGAAUCUACAUAUGUGUGCCUGCACGUACACA--UGCGUCGGUGUCUGGUGGCAGUCGUCUCAUGUGCCACAUAUGUA -(((((.(((((..((((.(((......))))))).))))).))))).((((((((((((....)))...((((--((.(.(((((((....)))).))).).))))))..))))))))) ( -33.40) >DroSim_CAF1 83 102 - 1 AGACCCCACAAUCGCUGUGUAUACAUUAAUGAAAGCAAUA---GGAUCUGUAUGG---------UAGG--CAUGUGUGUAUUGGGGUCUGGUGGCAA---UCUCAUGUGCCACACAUGU- ((((((((....((((((((.(((......((........---...))......)---------)).)--)))).)))...)))))))).((((((.---.......))))))......- ( -30.60) >DroYak_CAF1 83 100 - 1 AGGCCCCACAAUCGCUGUGUAUACAUUAAUGAAAGCAAUA---GGAUCUAUAUGG---------AAGG--CAUG--UGUAUUGGGGUCUGGUGGCAA---UCUCAUGUGCCACACAUGU- (((((((......(((...(((......)))..)))....---.(((.((((((.---------....--))))--)).)))))))))).((((((.---.......))))))......- ( -29.70) >DroPer_CAF1 5872 117 - 1 -GUUCCCAUAUUCACUUUGUAUAUAUUAAUGAAAGCAAUAUAGGAAUCUACAUAUGUGUUCCUGCACGUACACA--UGUGUUGGUGUCUGGUGGCAGUCGUCUCAUGUGCCACAUAUGUA -(((((.(((((..((((.(((......))))))).))))).))))).....(((((((....))))))).(((--(((((.((..(.(((.(((....)))))).)..)))))))))). ( -30.70) >consensus AGACCCCACAAUCGCUGUGUAUACAUUAAUGAAAGCAAUA___GGAUCUAUAUGG_________AACG__CAUG__UGUAUUGGGGUCUGGUGGCAA___UCUCAUGUGCCACACAUGU_ .((((((....(((..(((....)))...)))..(((.......................................)))...))))))..((((((...........))))))....... (-13.97 = -14.21 + 0.24)

| Location | 18,151,706 – 18,151,799 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.38 |
| Mean single sequence MFE | -22.22 |
| Consensus MFE | -5.98 |
| Energy contribution | -5.82 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.27 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18151706 93 - 20766785 ACUGAAGAGACGAUCUCAUAUCGUUCAAAUGUGGGGAAUCGUAUAUGUACAU--AUAUA------UGAAG----------ACGUACAUAUG----UGCAAUUGUA-----CCCCUUUCAC ..(((((.((((((.....)))))).......((((...((....(((((((--((.((------((...----------.)))).)))))----))))..))..-----))))))))). ( -24.10) >DroPse_CAF1 5902 99 - 1 ACUCGAGAGUCGAUCUCAUAUCGCUCAACUUGAAGGCAUCCUAUGCCU-CUU--CUAUU------UGAGU----------AUCUGCCUCUGUUAUUGCAAUUGUAUUCCACCCCAUUC-- (((((((((.((((.....))))))).......((((((...))))))-...--....)------)))))----------...(((...(((....)))...))).............-- ( -18.30) >DroSim_CAF1 185 88 - 1 ACUGAAGAGACGAUCUCAUAUCGCUCAAAUGUGGGGAAUCGUAUAUGU------AUAUA------AGAAU----------AC-UACAUAUG----UGCAAUUGUA-----CCCCUUUCAC ..(((((((.((((.....)))))))......((((.....(((((((------((((.------...))----------).-))))))))----(((....)))-----)))).)))). ( -20.70) >DroYak_CAF1 183 111 - 1 ACUCAAGAGACGAUCUCAUAUCGCUCAAAUGGGGGGAAUCAUAUACAUAUGUGUAUAUACAAAUAAGAACAUACAUAUGUAUGUACAUAUG----UGCAAUUGUA-----CCCCUUUCAC ......(((.((((.....)))))))....((((((...((....((((((((((((((((................))))))))))))))----))....))..-----)))))).... ( -32.79) >DroPer_CAF1 5989 99 - 1 ACUCGAGAGUCGAUCUCAUAUCGCUCAACUUGAAGGAAUCGUAUGCCU-CUU--CUAUA------UGAGU----------AUCUGCCUCUGUUAUUGCAAUUGUAUUCCACCCCAUUC-- ......(((.((((.....)))))))........(((((((((((...-...--.))))------)))((----------(...((....))...))).......)))).........-- ( -15.20) >consensus ACUCAAGAGACGAUCUCAUAUCGCUCAAAUGGGGGGAAUCGUAUACCU_CUU__AUAUA______UGAAU__________AUCUACAUAUG____UGCAAUUGUA_____CCCCUUUCAC ......(((.((((.....))))))).............................................................................................. ( -5.98 = -5.82 + -0.16)

| Location | 18,151,737 – 18,151,839 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.38 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -8.84 |
| Energy contribution | -9.92 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.35 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18151737 102 - 20766785 ACGUCCCCCAAUCUUUUGUUUAGAGCGAGAGAAACCAAGAACUGAAGAGACGAUCUCAUAUCGUUCAAAUGUGGGGAAUCGUAUAUGUACAU--AUAUA------UGAAG---------- ...(((((((.((((((((.....))))))))......((((.((.(((.....)))...))))))...)).))))).(((((((((....)--)))))------)))..---------- ( -29.90) >DroPse_CAF1 5940 86 - 1 G-----------CUUCAUUUU--AGC--GAGAAACAAACGACUCGAGAGUCGAUCUCAUAUCGCUCAACUUGAAGGCAUCCUAUGCCU-CUU--CUAUU------UGAGU---------- (-----------((.......--)))--((((......(((((....))))).)))).....((((((...((((((((...))))))-..)--)...)------)))))---------- ( -22.50) >DroSim_CAF1 215 98 - 1 ACGUCCCCCAAUCUUUUGUUUACAGCGAGAGAAACCAAGAACUGAAGAGACGAUCUCAUAUCGCUCAAAUGUGGGGAAUCGUAUAUGU------AUAUA------AGAAU---------- ((((((((((.((((((((.....))))))))..............(((.((((.....)))))))...)).)))))..)))......------.....------.....---------- ( -24.90) >DroYak_CAF1 214 119 - 1 ACUUC-CCCAAUCUUUUGUUUAGAGCGAGAGAAACCAACAACUCAAGAGACGAUCUCAUAUCGCUCAAAUGGGGGGAAUCAUAUACAUAUGUGUAUAUACAAAUAAGAACAUACAUAUGU ..(((-(((..((((((((.....))))))))..............(((.((((.....)))))))......))))))......((((((((((................)))))))))) ( -31.49) >DroPer_CAF1 6027 90 - 1 G-------GGGUCUUCAUUUU--AAC--GAGAAACAAACGACUCGAGAGUCGAUCUCAUAUCGCUCAACUUGAAGGAAUCGUAUGCCU-CUU--CUAUA------UGAGU---------- (-------(..((((((....--..(--(((..........)))).(((.((((.....)))))))....))))))..))......((-(..--.....------.))).---------- ( -18.80) >consensus AC_UC_CCCAAUCUUUUGUUUA_AGCGAGAGAAACCAACAACUCAAGAGACGAUCUCAUAUCGCUCAAAUGGGGGGAAUCGUAUACCU_CUU__AUAUA______UGAAU__________ ...........((((((((.....))))))))..............(((.((((.....)))))))...................................................... ( -8.84 = -9.92 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:49 2006