| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,116,125 – 18,116,225 |
| Length | 100 |
| Max. P | 0.708377 |

| Location | 18,116,125 – 18,116,225 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -26.19 |
| Energy contribution | -26.13 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
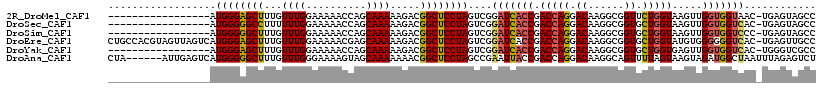
>2R_DroMel_CAF1 18116125 100 + 20766785 -----------------AUGGGAGCUUUGUUUGGAAAAACCAGCAAAAAGACGGCUCCUAGUCGGAUCACCGACCAGGACAAGGCGGUUCUGGUAAGUUGGUGGUAAC-UGAGUAGCC -----------------.(((((((((((((.((.....)))))))......)))))))).(((((((((((((((((((......))))))))....)))))))..)-)))...... ( -35.90) >DroSec_CAF1 1 100 + 1 -----------------AUGGGGGCCUUUUUUGGAAAAACCAGCAAAAAGACGGCUCCUAGUCGGAUCACCGACCAGGACAAGGCGGUGCUGGUAAGUUGGUGGUCAC-UGAGUAGCC -----------------.((((((((((((((((.....)))....))))).))))))))....(((((((((((((.((......)).)))))....))))))))..-......... ( -35.90) >DroSim_CAF1 1 100 + 1 -----------------AUGGGGGCUUUGUUUGGAAAAACCAGCAAAAAGACGGCUCCUAGUCGGAUCACCGACCAGGACAAGGCGGUGCUGGUAAGUUGGUGGUCCC-UGAGUAGCC -----------------..(((((((....(..(....(((((((......((.(((((.(((((....))))).))))...).)).)))))))...)..).))))))-)........ ( -37.10) >DroEre_CAF1 685 117 + 1 CUGCCACGUAGUUAGUCAUGGGAGCUUUGUUUGGAAAAACGAGCAAAAAGACGGCUCCUAGUCGGAUCACCGACCAGGACAAGGCGGUGCUGGUAUGUGGGGGGUCAC-UGAGUUGCC ((.(((((((........(((((((((((((((......)))))))......))))))))(((((....)))))(((.((......)).))).))))))).)).....-......... ( -40.80) >DroYak_CAF1 1 100 + 1 -----------------AUGGGAGCUUUGUUUGGAAAAACCAGCAAAAAGACGGCUCCUAGUCGGAUCACCGACCAGGACAAGGCGGUGCUGGUGAGUUGGUGGUCAC-UGGGUCGCC -----------------......(((..((((....)))).)))........(((.((((((..(((((((((((((.((......)).)))))....))))))))))-))))..))) ( -37.00) >DroAna_CAF1 203 112 + 1 CUA------AUUGAGUCAUGGGGGCUUUGUUUGGGAAAAGUAGCAAAAAAACGGCUCCUAGCCGAAUUACCGACCAGGACAAGGCAGUUUUAGUAAGUAGAUGGCUAAUUUAGAGUCU ...------...((.((.((((((((.(.((((..........))))...).))))))))((((..((((..((.(((((......))))).))..)))).)))).......)).)). ( -25.60) >consensus _________________AUGGGAGCUUUGUUUGGAAAAACCAGCAAAAAGACGGCUCCUAGUCGGAUCACCGACCAGGACAAGGCGGUGCUGGUAAGUUGGUGGUCAC_UGAGUAGCC ..................((((((((...((((..........)))).....))))))))....(((((((.(((((.((......)).))))).....)))))))............ (-26.19 = -26.13 + -0.05)
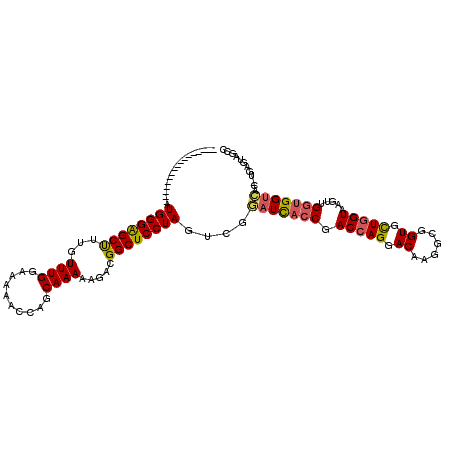

| Location | 18,116,125 – 18,116,225 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -28.99 |
| Consensus MFE | -21.88 |
| Energy contribution | -22.22 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18116125 100 - 20766785 GGCUACUCA-GUUACCACCAACUUACCAGAACCGCCUUGUCCUGGUCGGUGAUCCGACUAGGAGCCGUCUUUUUGCUGGUUUUUCCAAACAAAGCUCCCAU----------------- ((((....(-(((......)))).(((((.((.((....((((((((((....)))))))))))).)).......)))))............)))).....----------------- ( -25.90) >DroSec_CAF1 1 100 - 1 GGCUACUCA-GUGACCACCAACUUACCAGCACCGCCUUGUCCUGGUCGGUGAUCCGACUAGGAGCCGUCUUUUUGCUGGUUUUUCCAAAAAAGGCCCCCAU----------------- ((((....(-((........))).(((((((......((((((((((((....))))))))))..))......)))))))............)))).....----------------- ( -32.30) >DroSim_CAF1 1 100 - 1 GGCUACUCA-GGGACCACCAACUUACCAGCACCGCCUUGUCCUGGUCGGUGAUCCGACUAGGAGCCGUCUUUUUGCUGGUUUUUCCAAACAAAGCCCCCAU----------------- ((((.....-((.....)).....(((((((......((((((((((((....))))))))))..))......)))))))............)))).....----------------- ( -34.30) >DroEre_CAF1 685 117 - 1 GGCAACUCA-GUGACCCCCCACAUACCAGCACCGCCUUGUCCUGGUCGGUGAUCCGACUAGGAGCCGUCUUUUUGCUCGUUUUUCCAAACAAAGCUCCCAUGACUAACUACGUGGCAG (((......-(((......))).................((((((((((....)))))))))))))........(((.((((....))))..)))..(((((........)))))... ( -32.10) >DroYak_CAF1 1 100 - 1 GGCGACCCA-GUGACCACCAACUCACCAGCACCGCCUUGUCCUGGUCGGUGAUCCGACUAGGAGCCGUCUUUUUGCUGGUUUUUCCAAACAAAGCUCCCAU----------------- (((.....(-((........))).(((((((......((((((((((((....))))))))))..))......))))))).............))).....----------------- ( -29.30) >DroAna_CAF1 203 112 - 1 AGACUCUAAAUUAGCCAUCUACUUACUAAAACUGCCUUGUCCUGGUCGGUAAUUCGGCUAGGAGCCGUUUUUUUGCUACUUUUCCCAAACAAAGCCCCCAUGACUCAAU------UAG .((.((.....((((............(((((.((....((((((((((....)))))))))))).)))))...))))((((........)))).......)).))...------... ( -20.06) >consensus GGCUACUCA_GUGACCACCAACUUACCAGCACCGCCUUGUCCUGGUCGGUGAUCCGACUAGGAGCCGUCUUUUUGCUGGUUUUUCCAAACAAAGCCCCCAU_________________ ........................(((((((......((((((((((((....))))))))))..))......)))))))...................................... (-21.88 = -22.22 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:38 2006