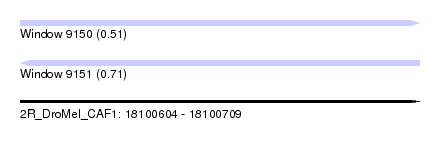
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,100,604 – 18,100,709 |
| Length | 105 |
| Max. P | 0.712348 |

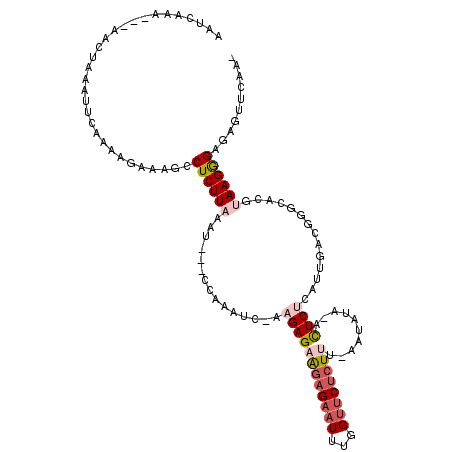
| Location | 18,100,604 – 18,100,709 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 69.45 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -4.31 |
| Energy contribution | -5.55 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.18 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
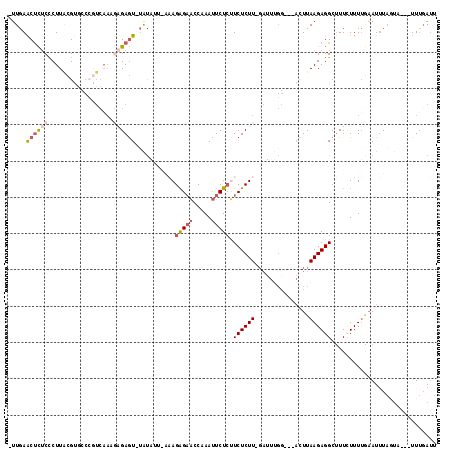
>2R_DroMel_CAF1 18100604 105 + 20766785 -UUGAACUCUCCCUUACGUGCCCAUCAAAGAGAGU-UAUAUUAAAAGAGAACCAAAUUCUCUUCUCUU-GAUUUGG---AUUUAAGAGGCUUUCUUUUGUAUUU-------UUAUAUU -......................((.(((((((((-........(((((((.....)))))))(((((-((.....---..)))))))))))))))).))....-------....... ( -20.00) >DroSec_CAF1 51841 109 + 1 -UUGAACUCUCCCUUACGUGCCCGUCAAUGAGAGU-UAUAUUGAAAGAGAACCAAAUUCUCUUCUCUU-GAUUUGG---ACUUAAGAGGCUUUAUUUUGAAUUUAGUA---UUUGAUU -...(((((((....(((....)))....))))))-)((((((((((((((.....))))))((((((-((.....---..))))))))............)))))))---)...... ( -23.30) >DroSim_CAF1 52595 109 + 1 -UUGAACUCUCCCUUACGUGCCCGUCAAUGAGAGU-UAUAUUGAAAGAGAACCAAAUUCUCUUCUCUU-GAUUUGG---ACUUAAGAGGCUUUAUUUUGAAUUUAGUA---UUUGAUU -...(((((((....(((....)))....))))))-)((((((((((((((.....))))))((((((-((.....---..))))))))............)))))))---)...... ( -23.30) >DroEre_CAF1 52529 100 + 1 CUUGAACCUACUCUUUGG-------------GAAU-UGCU----UAGAGAGCCCAAGUCUCCUCUCUUUUUGUCUCUUUAUUUAAGAGGCGUUCUUUUCAAUGUAGUUGAAUUUGAUU .......((((...(((.-------------.((.-.((.----.((((((..........))))))....(((((((.....)))))))))..))..))).))))............ ( -23.20) >DroYak_CAF1 54164 106 + 1 CUUAAGCUUUCUCUUGCGCACCAGUCAAA--GAGUCAGUU----CAGGGCACCAAAUUCUGCUCUCUUUGAAUAGG---CGUUAAGAGGCUUUCUUUGAAAUUUACUU---UUUGAGU ...((((...(((((((((.....(((((--(((.(((..----...(....).....)))..))))))))....)---)).))))))))))..((..(((......)---))..)). ( -28.20) >consensus _UUGAACUCUCCCUUACGUGCCCGUCAAAGAGAGU_UAUAUU_AAAGAGAACCAAAUUCUCUUCUCUU_GAUUUGG___ACUUAAGAGGCUUUCUUUUGAAUUUAGUA___UUUGAUU .....((((((..................))))))...........(((((.....))))).((((((...............))))))............................. ( -4.31 = -5.55 + 1.24)

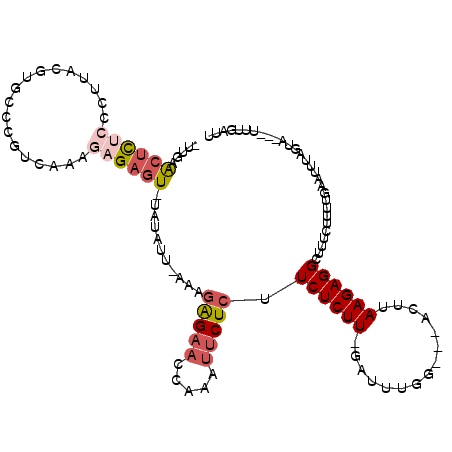
| Location | 18,100,604 – 18,100,709 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.45 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -5.79 |
| Energy contribution | -7.15 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.26 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18100604 105 - 20766785 AAUAUAA-------AAAUACAAAAGAAAGCCUCUUAAAU---CCAAAUC-AAGAGAAGAGAAUUUGGUUCUCUUUUAAUAUA-ACUCUCUUUGAUGGGCACGUAAGGGAGAGUUCAA- .......-------......((((((.((((((((...(---(......-..))...))))....)))).)))))).....(-(((((((((.(((....))).))))))))))...- ( -22.60) >DroSec_CAF1 51841 109 - 1 AAUCAAA---UACUAAAUUCAAAAUAAAGCCUCUUAAGU---CCAAAUC-AAGAGAAGAGAAUUUGGUUCUCUUUCAAUAUA-ACUCUCAUUGACGGGCACGUAAGGGAGAGUUCAA- .......---....................(((((....---.......-)))))((((((((...)))))))).......(-((((((....(((....)))....)))))))...- ( -21.10) >DroSim_CAF1 52595 109 - 1 AAUCAAA---UACUAAAUUCAAAAUAAAGCCUCUUAAGU---CCAAAUC-AAGAGAAGAGAAUUUGGUUCUCUUUCAAUAUA-ACUCUCAUUGACGGGCACGUAAGGGAGAGUUCAA- .......---....................(((((....---.......-)))))((((((((...)))))))).......(-((((((....(((....)))....)))))))...- ( -21.10) >DroEre_CAF1 52529 100 - 1 AAUCAAAUUCAACUACAUUGAAAAGAACGCCUCUUAAAUAAAGAGACAAAAAGAGAGGAGACUUGGGCUCUCUA----AGCA-AUUC-------------CCAAAGAGUAGGUUCAAG .......(((((.....)))))..(((((((((((.....)))))......((((((..........)))))).----.)).-((((-------------.....))))..))))... ( -18.30) >DroYak_CAF1 54164 106 - 1 ACUCAAA---AAGUAAAUUUCAAAGAAAGCCUCUUAACG---CCUAUUCAAAGAGAGCAGAAUUUGGUGCCCUG----AACUGACUC--UUUGACUGGUGCGCAAGAGAAAGCUUAAG .......---................(((((((((..((---((((.((((((((..(((..((..(....)..----))))).)))--))))).))).))).)))))...))))... ( -26.80) >consensus AAUCAAA___AACUAAAUUCAAAAGAAAGCCUCUUAAAU___CCAAAUC_AAGAGAAGAGAAUUUGGUUCUCUUU_AAUAUA_ACUCUCAUUGACGGGCACGUAAGGGAGAGUUCAA_ ..............................((((((...............((((((((((((...))))))))..........))))..............)))))).......... ( -5.79 = -7.15 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:30 2006