| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,082,883 – 18,082,988 |
| Length | 105 |
| Max. P | 0.896222 |

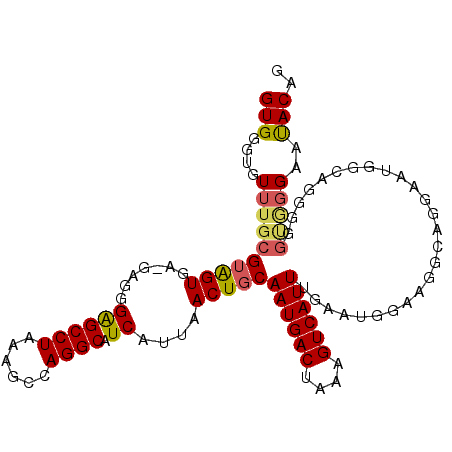
| Location | 18,082,883 – 18,082,988 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.71 |
| Mean single sequence MFE | -16.80 |
| Consensus MFE | -10.86 |
| Energy contribution | -10.75 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18082883 105 + 20766785 CUGUAUUCGCACCCCCUGCCAUUCCUGCCUUCCAUUCAAAUGACUUUAGUCAUUGCAGUUAAUGAUGCCUGGCUUUAGGCUCCCAC-UCACUACGCAAACACCCAC ........((.........((((.((((..........((((((....))))))))))..))))..(((((....)))))......-.......)).......... ( -17.70) >DroSec_CAF1 40427 105 + 1 CUGUAUUCCCACCCCCUGCCAUUCCUGCCUUCCAUUCAAAUGACUUUAGUCAUUGCAGUUAAUGAUGCCUGGCUUUAGGCUCCCUC-UCACUACGCAAACACCCAC ................(((((((.((((..........((((((....))))))))))..))))..(((((....)))))......-.......)))......... ( -17.30) >DroSim_CAF1 41080 105 + 1 CUGUAUUCCCACCCCCUGCCAUUCCUGCCUUCCAUUCAAAUGACUUUAGUCAUUGCAGUUAAUGAUGCCUGGCUUUAGGCUCCCUC-UCACUACGCAAACACCCAC ................(((((((.((((..........((((((....))))))))))..))))..(((((....)))))......-.......)))......... ( -17.30) >DroEre_CAF1 40880 104 + 1 CUGUAUUCCUGCCACCCGCCAUUC-UUCCUUCCAUUCAAAUGACUUUAGUCAUUGCAGUUAAUGAUGCCUGGCUUUAGGCUCACUC-CCACUACGCAAACACCCAC ..........(((....((((...-.............((((((....))))))(((........))).))))....)))......-................... ( -17.10) >DroYak_CAF1 41902 82 + 1 CUGUAUUCCCGCC------------------------AAAUGACUUUAGUCAUUGCAGUUAAUGAUGCCUGGCUUUAGGCUCCCCCGUCACUACGCAAAUACCCAC ..(((((..((((------------------------(((((((....))))))(((........))).))))....(((......))).....)..))))).... ( -17.40) >DroAna_CAF1 39758 82 + 1 CAGUG------------------UAUCCUUUCUAUUCAAAUGACUUUAGUCAUUGCAGUUAAUGAUGCCUGGCUUUAGGCCCCACC-AUACCUCCCA-----CCAC .((.(------------------((((.((...(((..((((((....))))))..))).)).)))))))(((.....))).....-..........-----.... ( -14.00) >consensus CUGUAUUCCCACCCCCUGCCAUUCCUGCCUUCCAUUCAAAUGACUUUAGUCAUUGCAGUUAAUGAUGCCUGGCUUUAGGCUCCCUC_UCACUACGCAAACACCCAC ..(((.................................((((((....))))))............(((((....)))))...........)))............ (-10.86 = -10.75 + -0.11)

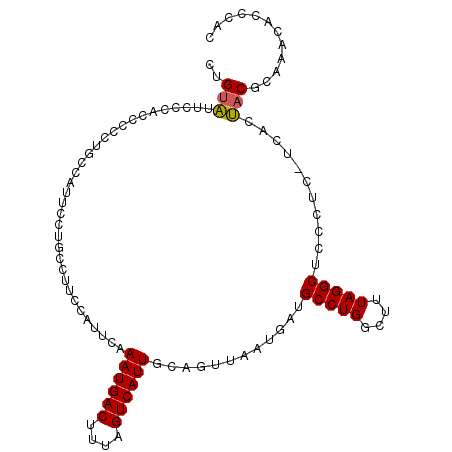
| Location | 18,082,883 – 18,082,988 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.71 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -17.49 |
| Energy contribution | -17.72 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18082883 105 - 20766785 GUGGGUGUUUGCGUAGUGA-GUGGGAGCCUAAAGCCAGGCAUCAUUAACUGCAAUGACUAAAGUCAUUUGAAUGGAAGGCAGGAAUGGCAGGGGGUGCGAAUACAG ....(((((((((((((.(-((((..((((......)))).))))).)))))....(((...(((((((...((.....)).)))))))....))))))))))).. ( -32.90) >DroSec_CAF1 40427 105 - 1 GUGGGUGUUUGCGUAGUGA-GAGGGAGCCUAAAGCCAGGCAUCAUUAACUGCAAUGACUAAAGUCAUUUGAAUGGAAGGCAGGAAUGGCAGGGGGUGGGAAUACAG ....((((((.((((((..-...(..((((......))))..)....)))))....(((...(((((((...((.....)).)))))))....)))).)))))).. ( -26.10) >DroSim_CAF1 41080 105 - 1 GUGGGUGUUUGCGUAGUGA-GAGGGAGCCUAAAGCCAGGCAUCAUUAACUGCAAUGACUAAAGUCAUUUGAAUGGAAGGCAGGAAUGGCAGGGGGUGGGAAUACAG ....((((((.((((((..-...(..((((......))))..)....)))))....(((...(((((((...((.....)).)))))))....)))).)))))).. ( -26.10) >DroEre_CAF1 40880 104 - 1 GUGGGUGUUUGCGUAGUGG-GAGUGAGCCUAAAGCCAGGCAUCAUUAACUGCAAUGACUAAAGUCAUUUGAAUGGAAGGAA-GAAUGGCGGGUGGCAGGAAUACAG (((....((((((((((..-.(((((((((......)))).))))).)))))....(((...(((((((............-))))))).))).)))))..))).. ( -30.00) >DroYak_CAF1 41902 82 - 1 GUGGGUAUUUGCGUAGUGACGGGGGAGCCUAAAGCCAGGCAUCAUUAACUGCAAUGACUAAAGUCAUUU------------------------GGCGGGAAUACAG ....((((((.(((.(((((...((.((((......)))).(((((......)))))))...)))))..------------------------.))).)))))).. ( -25.40) >DroAna_CAF1 39758 82 - 1 GUGG-----UGGGAGGUAU-GGUGGGGCCUAAAGCCAGGCAUCAUUAACUGCAAUGACUAAAGUCAUUUGAAUAGAAAGGAUA------------------CACUG ..((-----(((.((..((-((((.(((.....)))...))))))...)).)((((((....))))))...............------------------)))). ( -21.00) >consensus GUGGGUGUUUGCGUAGUGA_GAGGGAGCCUAAAGCCAGGCAUCAUUAACUGCAAUGACUAAAGUCAUUUGAAUGGAAGGCAGGAAUGGCAGGGGGUGGGAAUACAG (((....((((((((((.......((((((......)))).))....)))))((((((....))))))..........................)))))..))).. (-17.49 = -17.72 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:27 2006