| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,082,239 – 18,082,369 |
| Length | 130 |
| Max. P | 0.993993 |

| Location | 18,082,239 – 18,082,342 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 62.19 |
| Mean single sequence MFE | -25.56 |
| Consensus MFE | -7.94 |
| Energy contribution | -9.18 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18082239 103 + 20766785 AUAUUUUUUAUAUAUAUUGUAGGUUGGUUG-GUAUACGUACAUAUGUAUACGUACUGUAUGUACCGUAUAUAC--CGAUGCCUGUUGCCAGGUGCCAACUUGGCUC ...................(((((..((((-(((((.(((((((((((....))).))))))))....)))))--)))))))))..(((((((....))))))).. ( -33.10) >DroSec_CAF1 39813 84 + 1 AUAUCUAUAAUAUAUA---------GGAUG-GUGUACGUACGUAC------GUCCCGUAUG--CAG--UAUAC--CGAUGACUGUGACCAGGUGCCAACUUAGCUC ............((((---------(..((-((((((...(((((------(...))))))--..)--)))))--))....)))))...((((....))))..... ( -19.30) >DroSim_CAF1 40467 84 + 1 AUAUUUUUUAUAUAUA---------GGUUG-GUGUACGUACGUAU------GUACCGUAUA--CCG--UAUAC--CGAUGCCUGUUGCCAGGUGCCAACUUGGCUC .............(((---------(((((-(((((((((((...------....))))..--.))--)))))--))..)))))).(((((((....))))))).. ( -29.50) >DroYak_CAF1 41231 79 + 1 AUAUUUUGU---------------------UUUAUACUUGUAUUUAGGUAGG---AGUAUGUA-UGUAUAUGC--CGAUGCCUGUUGCCAGGUGCCAACUUGGCUC .........---------------------..............((((((..---.((((((.-...))))))--...))))))..(((((((....))))))).. ( -21.20) >DroAna_CAF1 39013 82 + 1 AUAUUUUUUAUAU----------------------ACCUACAUAUAUAGAGGUA-UGUGAGUG-AGUGGGGCGGGGAAGGCCUGUUGCCAGGUGCCAGGUUGGCUC ......(((((((----------------------((((.(.......))))))-)))))).(-(((.((.(..((...(((((....))))).)).).)).)))) ( -24.70) >consensus AUAUUUUUUAUAUAUA_________GG_UG_GUAUACGUACAUAU____A_GUACCGUAUGU_CAGU_UAUAC__CGAUGCCUGUUGCCAGGUGCCAACUUGGCUC ...................................((((((((((......))).))))))).................(((((....)))))(((.....))).. ( -7.94 = -9.18 + 1.24)

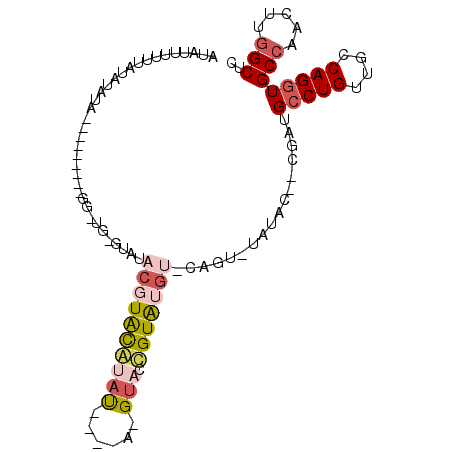
| Location | 18,082,239 – 18,082,342 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 62.19 |
| Mean single sequence MFE | -19.64 |
| Consensus MFE | -10.52 |
| Energy contribution | -10.56 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18082239 103 - 20766785 GAGCCAAGUUGGCACCUGGCAACAGGCAUCG--GUAUAUACGGUACAUACAGUACGUAUACAUAUGUACGUAUAC-CAACCAACCUACAAUAUAUAUAAAAAAUAU .......(((((..((((....))))....(--((((((...(((((((..(((....))).)))))))))))))-)..)))))...................... ( -27.70) >DroSec_CAF1 39813 84 - 1 GAGCUAAGUUGGCACCUGGUCACAGUCAUCG--GUAUA--CUG--CAUACGGGAC------GUACGUACGUACAC-CAUCC---------UAUAUAUUAUAGAUAU (((((.....)))..(((....)))...))(--((.((--(((--(.((((...)------))).))).))).))-)...(---------((((...))))).... ( -15.50) >DroSim_CAF1 40467 84 - 1 GAGCCAAGUUGGCACCUGGCAACAGGCAUCG--GUAUA--CGG--UAUACGGUAC------AUACGUACGUACAC-CAACC---------UAUAUAUAAAAAAUAU (((((.....))).((((....))))..))(--((...--.((--(.((((.(((------....))))))).))-).)))---------................ ( -23.10) >DroYak_CAF1 41231 79 - 1 GAGCCAAGUUGGCACCUGGCAACAGGCAUCG--GCAUAUACA-UACAUACU---CCUACCUAAAUACAAGUAUAAA---------------------ACAAAAUAU .......(((((..((((....))))..)))--)).......-...(((((---..((......))..)))))...---------------------......... ( -14.20) >DroAna_CAF1 39013 82 - 1 GAGCCAACCUGGCACCUGGCAACAGGCCUUCCCCGCCCCACU-CACUCACA-UACCUCUAUAUAUGUAGGU----------------------AUAUAAAAAAUAU ..(((.....))).((((....))))................-.......(-((((((.......).))))----------------------))........... ( -17.70) >consensus GAGCCAAGUUGGCACCUGGCAACAGGCAUCG__GUAUA_ACGG_ACAUACAGUAC_U____AUAUGUACGUACAC_CA_CC_________UAUAUAUAAAAAAUAU ..(((.....))).((((....))))................................................................................ (-10.52 = -10.56 + 0.04)

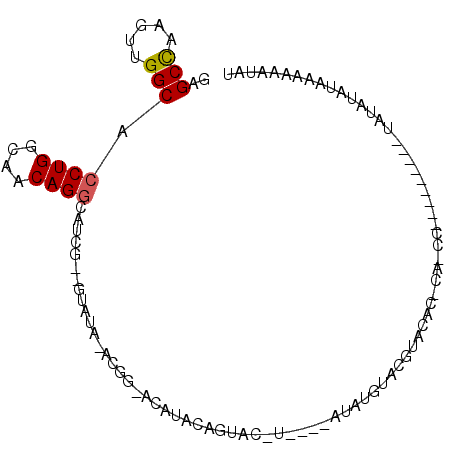
| Location | 18,082,276 – 18,082,369 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 74.40 |
| Mean single sequence MFE | -19.34 |
| Consensus MFE | -11.44 |
| Energy contribution | -11.36 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18082276 93 - 20766785 AAGAAAAUCAAUACGAGGCAGACACAAGAGCCAAGUUGGCACCUGGCAACAGGCAUCG--GUAUAUACGGUACAUACAGUACGUAUACAUAUGUA .............(((.((..........(((.....)))..(((....))))).)))--((((((...((((.....))))))))))....... ( -22.80) >DroSec_CAF1 39841 81 - 1 AAGAAAAUCAAUACGAGGCAGA--CUAGAGCUAAGUUGGCACCUGGUCACAGUCAUCG--GUAUA--CUG--CAUACGGGAC------GUACGUA .............((((((.((--((((.(((.....)))..))))))...))).)))--((((.--(((--....)))...------))))... ( -17.50) >DroSim_CAF1 40495 81 - 1 AAGAAAAUCAAUACGAGGCAGA--CAAGAGCCAAGUUGGCACCUGGCAACAGGCAUCG--GUAUA--CGG--UAUACGGUAC------AUACGUA .............(((.((...--.....(((.....)))..(((....))))).)))--((((.--((.--....))))))------....... ( -19.40) >DroYak_CAF1 41248 87 - 1 AAGAAAAUCAAUACGAGGCAGA--CAAGAGCCAAGUUGGCACCUGGCAACAGGCAUCG--GCAUAUACA-UACAUACU---CCUACCUAAAUACA ................(((...--.....)))..(((((..((((....))))..)))--)).......-........---.............. ( -17.50) >DroAna_CAF1 39029 91 - 1 AAUAAAAUCAAUACAAGGCAGA--CAAGAGCCAACCUGGCACCUGGCAACAGGCCUUCCCCGCCCCACU-CACUCACA-UACCUCUAUAUAUGUA ................(((.(.--.(((.(((.....))).((((....)))).)))..).))).....-.....(((-((........))))). ( -19.50) >consensus AAGAAAAUCAAUACGAGGCAGA__CAAGAGCCAAGUUGGCACCUGGCAACAGGCAUCG__GUAUA_ACGG_ACAUACAGUAC_U____AUAUGUA .............(((.((..........(((.....)))..(((....))))).)))..................................... (-11.44 = -11.36 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:25 2006