| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,071,677 – 18,071,868 |
| Length | 191 |
| Max. P | 0.768192 |

| Location | 18,071,677 – 18,071,776 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -43.02 |
| Consensus MFE | -21.40 |
| Energy contribution | -20.93 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
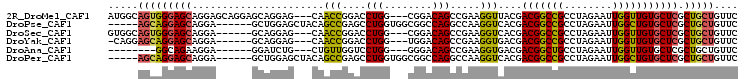
>2R_DroMel_CAF1 18071677 99 - 20766785 AUGGCAGUGGGAGCAGGAGCAGGAGCAGGAG---CAACCGGACCUGG---CGGACAGCCGAAGGUUACGACGGCCGCCUAGAAUUGGUUGUGCUCGCUGCUGUUC ..(((((..(((((....((....))....(---(((((((..((((---((....((((..(....)..)))))))).))..)))))))))))).)..))))). ( -40.70) >DroPse_CAF1 31718 94 - 1 -----AGCAGGAGCAGGA------GCUGGAGCUACAGCCGAGCCUGGUGGCGGCCAGGCCAAGGUCACGACGGCCGCCUAGAAUUGGCUGUGCUCGCUGCUGUUC -----.....((((((.(------((..((((.((((((((..(((..(((((((.(((....))).....))))))))))..))))))))))))))).)))))) ( -51.00) >DroSec_CAF1 29194 93 - 1 GUGGCAGUGGGAGCAGGA------GCAGGAG---CAACCGGACCUGG---CGGACAGCCGAAGGUCACGACGGCCGCCUAGAAUUGGUUGUGCUCGCUGCUGUUC (..((((((((.((....------))....(---(((((((..((((---((....((((..(....)..)))))))).))..)))))))).))))))))..).. ( -39.30) >DroYak_CAF1 29972 92 - 1 -CAGGAGCAGGAGCAGGA------GCAGGAG---CAACCGGACCUGG---UGGACAGCCGAAGGUGACGACGGCCGCCUAGAAUUGGUUGUGCUCGCUGCUGUUC -...((((((.(((..((------((....(---(((((((..((((---((....((((..(....)..)))))))).))..))))))))))))))).)))))) ( -41.40) >DroAna_CAF1 29384 85 - 1 --------GGCAGAAGGA------GGAUCUG---CUGUUGGUCCUGG---GGGACAGCCGAAGGUGACGACGGCUGCCUAGAAUUGGUUGUGCUCGCUGCUGUUC --------(((((..(((------(((((..---.....))))))..---(((.((((((..(....)..))))))))).............))..))))).... ( -34.70) >DroPer_CAF1 32135 94 - 1 -----AGCAGGAGCAGGA------GCUGGAGCUACAGCCGAGCCUGGUGGCGGCCAGGCCAAGGUCACGACGGCCGCCUAGAAUUGGCUGUGCUCGCUGCUGUUC -----.....((((((.(------((..((((.((((((((..(((..(((((((.(((....))).....))))))))))..))))))))))))))).)))))) ( -51.00) >consensus _____AGCAGGAGCAGGA______GCAGGAG___CAACCGGACCUGG___CGGACAGCCGAAGGUCACGACGGCCGCCUAGAAUUGGUUGUGCUCGCUGCUGUUC .....(((((((((......................(((....(((........))).....)))....(((((((........))))))))))).))))).... (-21.40 = -20.93 + -0.47)
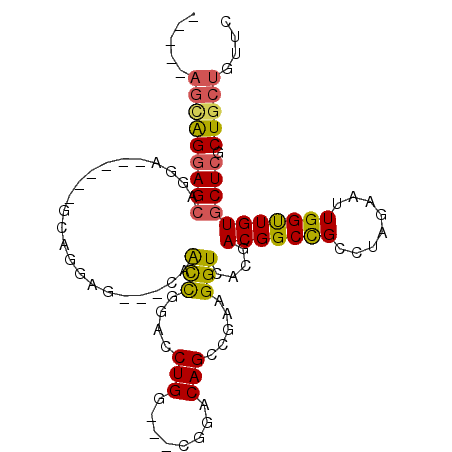
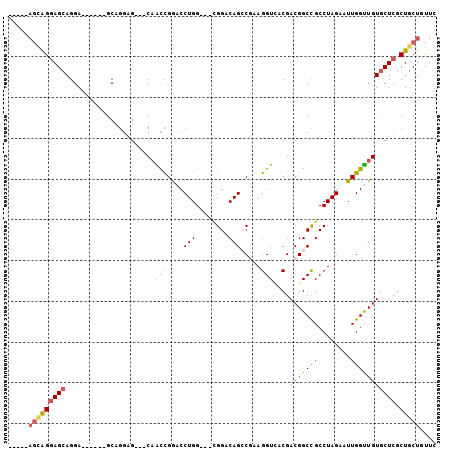
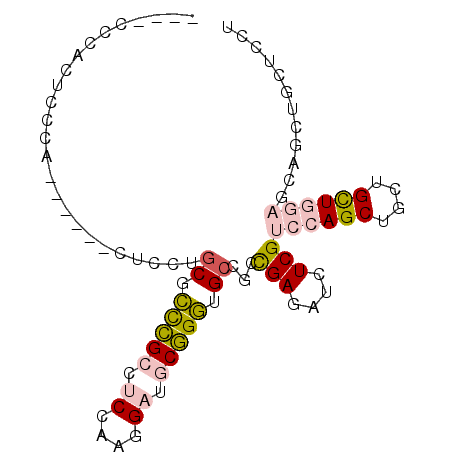
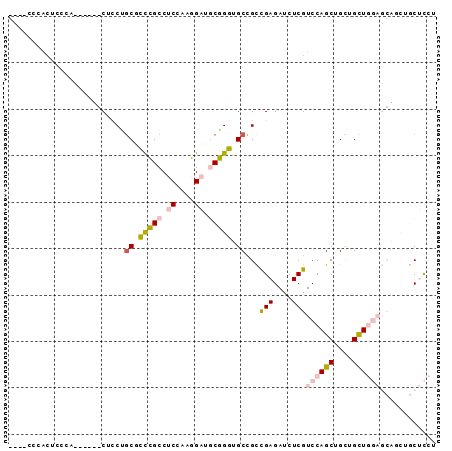
| Location | 18,071,776 – 18,071,868 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 65.10 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -15.48 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18071776 92 + 20766785 GCCGCCAACUCCCACUCCUGCAUCUGCGCCCGCCUCGAGGGAUGCGGGUGCCGCCGAGAUCUCGUCCAGCUGCUGUUGGAGCAGCUGCUCCU ..............(((..((((((((((((.......))).)))))))))....))).....(..((((((((.....))))))))..).. ( -34.70) >DroPse_CAF1 31812 73 + 1 ----CCCACUGCUC------CUCCAGCAGUGGCCUCAGCCGUCACUGUCGCCGUUGAGAUCUCGUCCAGCUGCAGCU---------CCGUGU ----.((((((((.------....))))))))..........(((.((.((.((((.(((...))))))).)).)).---------..))). ( -22.20) >DroSec_CAF1 29287 92 + 1 GCCGCCCACUCCCACUCCUGCGUCUGCGCCCGCCUCCAAGGAUGCGGGUGCCGCCGAGAUCUCGUCCAGCUGCUGCUGGAGCAUCUGCUCCU ...((.........(((..(((...((((((((.((....)).))))))))))).)))......((((((....)))))))).......... ( -36.80) >DroSim_CAF1 29434 92 + 1 GCCGCCCACUACCACUCCUGCGUCUGCGCCCGCCUCCAAGGAUGCGGGUGCCGCCGAGAUCUCGUCCAGCUGCUGCUGGAGCAGCUGCUCCU ((.((.........(((..(((...((((((((.((....)).))))))))))).)))......((((((....)))))))).))....... ( -39.00) >DroYak_CAF1 30064 72 + 1 --------------------CUCCUACUCCCGCCUCCAGGGAGGCGGGUGCUGCCGAGAUCUCGUCCAGCUGUUGCUGGAGCAGCUGCUCCU --------------------........((((((((....)))))))).((((((((....)))((((((....)))))))))))....... ( -38.30) >DroPer_CAF1 32229 73 + 1 ----CCCACUGCUC------CUCCAGCAGUGGCCUCAGCCGUCACUGUCGCCGUUGAGAUCUCGUCCAGCUGCAGCU---------CCGUGU ----.((((((((.------....))))))))..........(((.((.((.((((.(((...))))))).)).)).---------..))). ( -22.20) >consensus ____CCCACUCCCA______CUCCUGCGCCCGCCUCCAAGGAUGCGGGUGCCGCCGAGAUCUCGUCCAGCUGCUGCUGGAGCAGCUGCUCCU .........................((.(((((.((....)).))))).))...(((....)))((((((....))))))............ (-15.48 = -15.85 + 0.37)

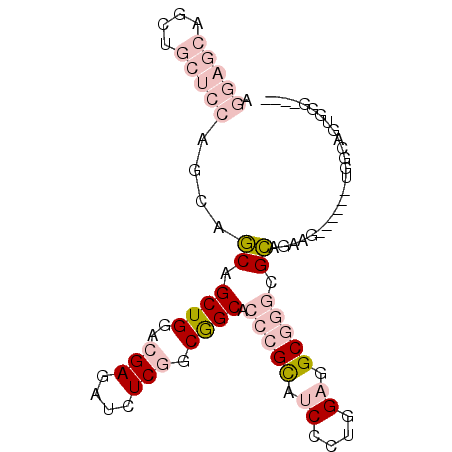
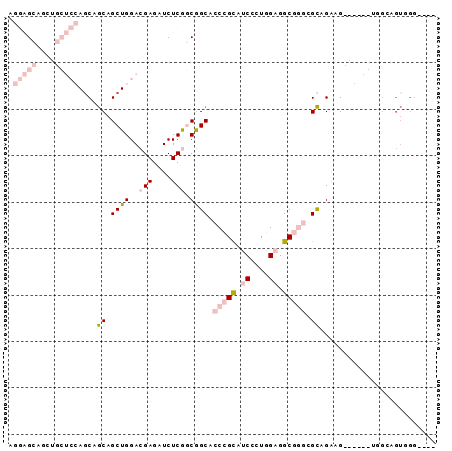
| Location | 18,071,776 – 18,071,868 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 65.10 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -15.85 |
| Energy contribution | -18.55 |
| Covariance contribution | 2.70 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18071776 92 - 20766785 AGGAGCAGCUGCUCCAACAGCAGCUGGACGAGAUCUCGGCGGCACCCGCAUCCCUCGAGGCGGGCGCAGAUGCAGGAGUGGGAGUUGGCGGC .(((((....)))))..((((.((((..(((....))).)))).(((((.(((((((...)))).((....)).)))))))).))))..... ( -37.10) >DroPse_CAF1 31812 73 - 1 ACACGG---------AGCUGCAGCUGGACGAGAUCUCAACGGCGACAGUGACGGCUGAGGCCACUGCUGGAG------GAGCAGUGGG---- ...((.---------.((((..((((...((....))..))))..))))..)).......((((((((....------.)))))))).---- ( -29.80) >DroSec_CAF1 29287 92 - 1 AGGAGCAGAUGCUCCAGCAGCAGCUGGACGAGAUCUCGGCGGCACCCGCAUCCUUGGAGGCGGGCGCAGACGCAGGAGUGGGAGUGGGCGGC .(((((....))))).((.((.((((..(((....))).)))).(((((.((((((...((....))...)).))))))))).))..))... ( -38.70) >DroSim_CAF1 29434 92 - 1 AGGAGCAGCUGCUCCAGCAGCAGCUGGACGAGAUCUCGGCGGCACCCGCAUCCUUGGAGGCGGGCGCAGACGCAGGAGUGGUAGUGGGCGGC ....((.(((((((((((....))))))......(((.(((((.(((((.((....)).))))).))...)))..)))..)))))..))... ( -39.90) >DroYak_CAF1 30064 72 - 1 AGGAGCAGCUGCUCCAGCAACAGCUGGACGAGAUCUCGGCAGCACCCGCCUCCCUGGAGGCGGGAGUAGGAG-------------------- ....((.(((((((((((....))))))(((....)))))))).((((((((....)))))))).)).....-------------------- ( -40.40) >DroPer_CAF1 32229 73 - 1 ACACGG---------AGCUGCAGCUGGACGAGAUCUCAACGGCGACAGUGACGGCUGAGGCCACUGCUGGAG------GAGCAGUGGG---- ...((.---------.((((..((((...((....))..))))..))))..)).......((((((((....------.)))))))).---- ( -29.80) >consensus AGGAGCAGCUGCUCCAGCAGCAGCUGGACGAGAUCUCGGCGGCACCCGCAUCCCUGGAGGCGGGCGCAGAAG______UGGCAGUGGG____ .(((((....)))))....((.((((..(((....))).)))).(((((.((....)).))))).))......................... (-15.85 = -18.55 + 2.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:22 2006