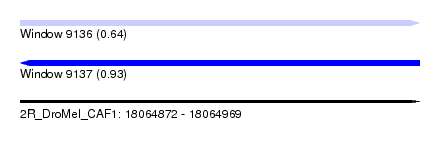
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,064,872 – 18,064,969 |
| Length | 97 |
| Max. P | 0.929085 |

| Location | 18,064,872 – 18,064,969 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 84.97 |
| Mean single sequence MFE | -22.59 |
| Consensus MFE | -15.71 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18064872 97 + 20766785 AAGUCGGCCACUCGAAAGAUCAGCGGAGUAUAUAUGUACAUAUGUACAUAGAGGCCUGAUAACGAGCGGAAUGGAAAGAAGGAAAAAUCGGCGGGGG ..(((((((.((((....(((((....((...(((((((....)))))))...)))))))..)))).))..(....)..........)))))..... ( -24.50) >DroSec_CAF1 22572 96 + 1 AAGUCGGCCACUCGAAAGAUCAGCAGAGUAUAUAUAUACAUAUGUACAUAGAGGCCUGAUAACGAGCGGAACGGAAAGUAAGAAAAAUCGGCGGG-G ..(((((((.((((....((((((...((((((((....))))))))......).)))))..)))).)).((.....))........)))))...-. ( -22.70) >DroSim_CAF1 22628 96 + 1 AAGUCGGCCACUCGAAAGAUCAGCAGAGUAUAUAUAUACAUAUGUACAUAGAGGCCUGAUAACGAGCGGAACGGAAAGAAAUGAAAAUCGGCGGG-G ..(((((((.((((....((((((...((((((((....))))))))......).)))))..)))).))..((........))....)))))...-. ( -21.70) >DroYak_CAF1 23154 84 + 1 AAGUCGGCCACUCGAAAGAUCAGCAGAGUAUACAGA-------GUAUACGGCCUCCUGAUAACGAGCGGAACGGAAAG----AAAAAUCGCUGGG-- .((..((((..((....))........((((((...-------))))))))))..)).....(.(((((.........----.....))))).).-- ( -21.44) >consensus AAGUCGGCCACUCGAAAGAUCAGCAGAGUAUAUAUAUACAUAUGUACAUAGAGGCCUGAUAACGAGCGGAACGGAAAGAAAGAAAAAUCGGCGGG_G ..(((((((.((((....(((((....((((((((....))))))))........)))))..)))).))..(.....).........)))))..... (-15.71 = -16.40 + 0.69)

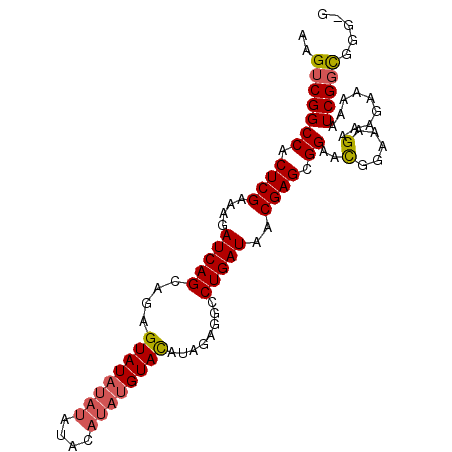
| Location | 18,064,872 – 18,064,969 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 84.97 |
| Mean single sequence MFE | -20.87 |
| Consensus MFE | -16.65 |
| Energy contribution | -16.71 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18064872 97 - 20766785 CCCCCGCCGAUUUUUCCUUCUUUCCAUUCCGCUCGUUAUCAGGCCUCUAUGUACAUAUGUACAUAUAUACUCCGCUGAUCUUUCGAGUGGCCGACUU ............................(((((((..((((((....(((((((....))))))).......).)))))....)))))))....... ( -20.40) >DroSec_CAF1 22572 96 - 1 C-CCCGCCGAUUUUUCUUACUUUCCGUUCCGCUCGUUAUCAGGCCUCUAUGUACAUAUGUAUAUAUAUACUCUGCUGAUCUUUCGAGUGGCCGACUU .-.......................((((((((((..(((((.....(((((((....))))))).........)))))....)))))))..))).. ( -19.44) >DroSim_CAF1 22628 96 - 1 C-CCCGCCGAUUUUCAUUUCUUUCCGUUCCGCUCGUUAUCAGGCCUCUAUGUACAUAUGUAUAUAUAUACUCUGCUGAUCUUUCGAGUGGCCGACUU .-.......................((((((((((..(((((.....(((((((....))))))).........)))))....)))))))..))).. ( -19.44) >DroYak_CAF1 23154 84 - 1 --CCCAGCGAUUUUU----CUUUCCGUUCCGCUCGUUAUCAGGAGGCCGUAUAC-------UCUGUAUACUCUGCUGAUCUUUCGAGUGGCCGACUU --.............----......((((((((((..(((((.(((..((((((-------...))))))))).)))))....)))))))..))).. ( -24.20) >consensus C_CCCGCCGAUUUUUCUUUCUUUCCGUUCCGCUCGUUAUCAGGCCUCUAUGUACAUAUGUAUAUAUAUACUCUGCUGAUCUUUCGAGUGGCCGACUU ............................(((((((..(((((.....(((((((....))))))).........)))))....)))))))....... (-16.65 = -16.71 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:17 2006