| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,062,521 – 18,062,616 |
| Length | 95 |
| Max. P | 0.921029 |

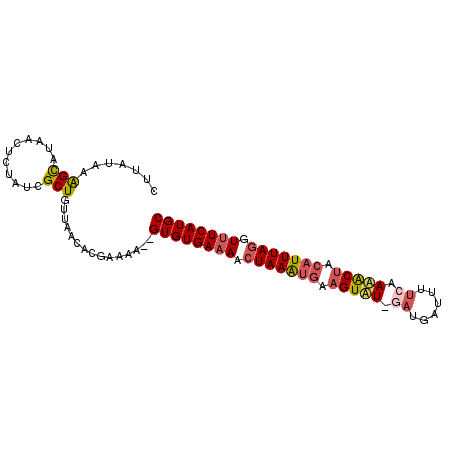
| Location | 18,062,521 – 18,062,616 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -20.95 |
| Consensus MFE | -13.51 |
| Energy contribution | -14.58 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18062521 95 + 20766785 CUUAUAAAGUAUAUCUCAAUCGCUGUUAACACAAUAAGAGUGUGAAAACUAAAUGAAGUAUCUAUGAUUUUCAAUACUUCAUUUAGGUUUCAUGC ....................((((.(((......))).))))(((((.(((((((((((((...((.....))))))))))))))).)))))... ( -22.50) >DroSim_CAF1 20214 93 + 1 CUUAUAAAGUAUAUCUCUAUCGCUGUUAACACGAUAA--GUGUGAAAACUAAAUGAAGUAUGGAUGAUUUUCAAUACUCCAUUUAGGUUUCAUGC .................(((((.((....))))))).--((((((((.(((((((.(((((.((......)).))))).))))))).)))))))) ( -24.40) >DroEre_CAF1 20400 91 + 1 UAUAUAAAGCAUAACUGCUUCGCUGUUAACUCGCAAA--GUGUGAAAAAUAAGUGAAGUAU--AUAAUUUACUAAGCAACAUUUAGGUUUCAUGC ........((((((((((((((((......((((...--..))))......))))))))).--........(((((.....))))))))..)))) ( -18.50) >DroYak_CAF1 20811 93 + 1 CUUAUGAGGCAUAACUGUUUCGCUGUUAACUCGCAAA--GUGUGAAAACUAAGCAGAGUUUAGAUGAUUUUCCAAGCUGCAUUUAGCUGUCAUGC .....((((((....)))))).(((..(((((((..(--((......)))..)).))))))))(((((......(((((....)))))))))).. ( -18.40) >consensus CUUAUAAAGCAUAACUCUAUCGCUGUUAACACGAAAA__GUGUGAAAACUAAAUGAAGUAU_GAUGAUUUUCAAAACUACAUUUAGGUUUCAUGC .......(((...........)))...............((((((((.(((((((.(((((.((......)).))))).))))))).)))))))) (-13.51 = -14.58 + 1.06)

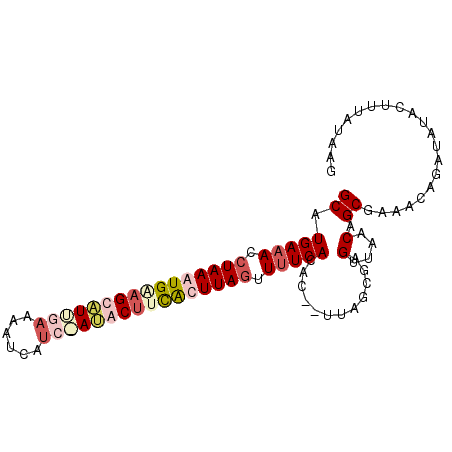
| Location | 18,062,521 – 18,062,616 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -11.49 |
| Energy contribution | -14.43 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18062521 95 - 20766785 GCAUGAAACCUAAAUGAAGUAUUGAAAAUCAUAGAUACUUCAUUUAGUUUUCACACUCUUAUUGUGUUAACAGCGAUUGAGAUAUACUUUAUAAG ...(((((.((((((((((((((..........)))))))))))))).)))))..(((..(((((.......))))).))).............. ( -24.80) >DroSim_CAF1 20214 93 - 1 GCAUGAAACCUAAAUGGAGUAUUGAAAAUCAUCCAUACUUCAUUUAGUUUUCACAC--UUAUCGUGUUAACAGCGAUAGAGAUAUACUUUAUAAG ((..((((.(((((((((((((.((......)).))))))))))))).))))((((--.....)))).....)).((((((.....))))))... ( -26.00) >DroEre_CAF1 20400 91 - 1 GCAUGAAACCUAAAUGUUGCUUAGUAAAUUAU--AUACUUCACUUAUUUUUCACAC--UUUGCGAGUUAACAGCGAAGCAGUUAUGCUUUAUAUA ((((((.....(((((......((((......--.)))).....)))))......(--(((((.........))))))...))))))........ ( -12.40) >DroYak_CAF1 20811 93 - 1 GCAUGACAGCUAAAUGCAGCUUGGAAAAUCAUCUAAACUCUGCUUAGUUUUCACAC--UUUGCGAGUUAACAGCGAAACAGUUAUGCCUCAUAAG (((((((.((.....)).(((((((......))))(((((.((..(((......))--)..)))))))...)))......)))))))........ ( -23.30) >consensus GCAUGAAACCUAAAUGAAGCAUUGAAAAUCAUC_AUACUUCACUUAGUUUUCACAC__UUAGCGAGUUAACAGCGAAACAGAUAUACUUUAUAAG ((.(((((.((((((((((((((((......)))))))))))))))).)))))............(....).))..................... (-11.49 = -14.43 + 2.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:14 2006