
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,046,844 – 18,046,978 |
| Length | 134 |
| Max. P | 0.995324 |

| Location | 18,046,844 – 18,046,949 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 93.27 |
| Mean single sequence MFE | -18.62 |
| Consensus MFE | -17.05 |
| Energy contribution | -17.55 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
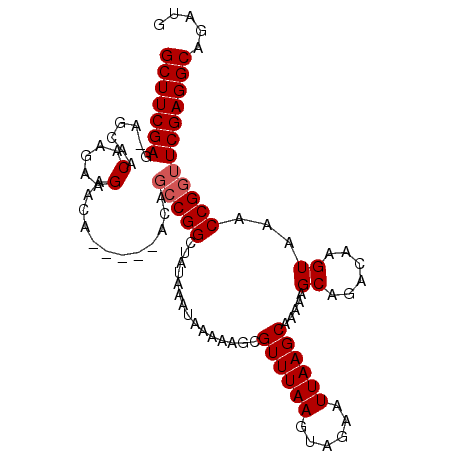
>2R_DroMel_CAF1 18046844 105 + 20766785 GCUUCGAA-UGCAACAAUAAGAACAACAAAAUAGCCGGCUAUAAAUAAAAAGCGUUUAAGUAGAAUUAAGCAAAAAGCAGACAAGUAAACCGGUUCGAGGCAGAUG ((((((((-(......))...............(((((...............((((((......)))))).....((......))...))))))))))))..... ( -19.20) >DroSec_CAF1 4351 103 + 1 GCUUCGAG-AGCAACAAGAAGAAUAACA--ACAGCCGGCUAUAAAUAAAAAGCGUUUAAGUAGAAUUAAGCAAAAAGCAGACAAGUAAACCGGUUCGAGGCAGAUG (((((((.-.....(.....).......--...(((((...............((((((......)))))).....((......))...))))))))))))..... ( -19.10) >DroEre_CAF1 4471 101 + 1 GCUUCGAGAAGCAACAAGAAGAACA-----ACAGCCGGCUAUAAAUAAAAAGCGUUUAAGUAGAAUUAAGCAAAAAGCAGACAAGUAAACCGGUUCGAGGCAGAUG ((((((((......)..........-----...(((((...............((((((......)))))).....((......))...))))))))))))..... ( -19.20) >DroYak_CAF1 4695 100 + 1 GCUUCGAG-AGCAACAAGAAGAACA-----ACACGCGGCUAUAAAUAAAAAGCGUUUAAGUAGAAUUAAGCAAAAAGCAGACAAGUAAACCGGUUCGAGGCAGAUG (((((((.-................-----....((.(((..........)))((((((......)))))).....))..((..(....)..)))))))))..... ( -17.00) >consensus GCUUCGAG_AGCAACAAGAAGAACA_____ACAGCCGGCUAUAAAUAAAAAGCGUUUAAGUAGAAUUAAGCAAAAAGCAGACAAGUAAACCGGUUCGAGGCAGAUG (((((((.......(.....)............(((((...............((((((......)))))).....((......))...))))))))))))..... (-17.05 = -17.55 + 0.50)

| Location | 18,046,882 – 18,046,978 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 98.35 |
| Mean single sequence MFE | -17.40 |
| Consensus MFE | -17.40 |
| Energy contribution | -17.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18046882 96 + 20766785 UAUAAAUAAAAAGCGUUUAAGUAGAAUUAAGCAAAAAGCAGACAAGUAAACCGGUUCGAGGCAGAUGCCUUAAGCUGACACAAA---AAAACCGACUCU ..............((((((......))))))...................(((((..((((....))))..))))).......---............ ( -17.40) >DroSec_CAF1 4387 96 + 1 UAUAAAUAAAAAGCGUUUAAGUAGAAUUAAGCAAAAAGCAGACAAGUAAACCGGUUCGAGGCAGAUGCCUUAAGCUGACACAAA---AAAACCGACUCU ..............((((((......))))))...................(((((..((((....))))..))))).......---............ ( -17.40) >DroSim_CAF1 4349 96 + 1 UAUAAAUAAAAAGCGUUUAAGUAGAAUUAAGCAAAAAGCAGACAAGUAAACCGGUUCGAGGCAGAUGCCUUAAGCUGACACAAA---AAAACCGACUCU ..............((((((......))))))...................(((((..((((....))))..))))).......---............ ( -17.40) >DroEre_CAF1 4505 95 + 1 UAUAAAUAAAAAGCGUUUAAGUAGAAUUAAGCAAAAAGCAGACAAGUAAACCGGUUCGAGGCAGAUGCCUUAAGCUGACACAAA----AAACCGACUCU ..............((((((......))))))...................(((((..((((....))))..))))).......----........... ( -17.40) >DroYak_CAF1 4728 99 + 1 UAUAAAUAAAAAGCGUUUAAGUAGAAUUAAGCAAAAAGCAGACAAGUAAACCGGUUCGAGGCAGAUGCCUUAAGCUGACACAAAAAAAAAACCGACUCU ..............((((((......))))))...................(((((..((((....))))..)))))...................... ( -17.40) >consensus UAUAAAUAAAAAGCGUUUAAGUAGAAUUAAGCAAAAAGCAGACAAGUAAACCGGUUCGAGGCAGAUGCCUUAAGCUGACACAAA___AAAACCGACUCU ..............((((((......))))))...................(((((..((((....))))..)))))...................... (-17.40 = -17.40 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:08 2006