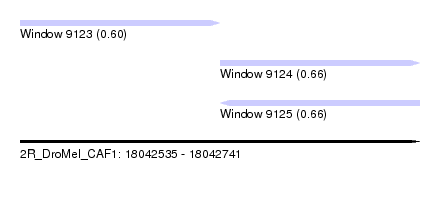
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,042,535 – 18,042,741 |
| Length | 206 |
| Max. P | 0.656487 |

| Location | 18,042,535 – 18,042,638 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.71 |
| Mean single sequence MFE | -35.59 |
| Consensus MFE | -24.08 |
| Energy contribution | -27.00 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18042535 103 + 20766785 CAGAUUGCAUUGUC----CAGCUACUGAAAUACACA-GAUACUCACAAUGCGAUCGGAGCGCUCGCCGCAGGGCAAGCGGGCCACUGCGGACUCGGGACUGUCCUCUG ..(((((((((((.----.((.(((((.......))-).)))).)))))))))))(((...(((((((((((((......))).))))))...))))....))).... ( -38.70) >DroVir_CAF1 155 88 + 1 ---U-----------------CUAUUGGCAUAAACAGUUUACUCACAAUACGAUCGGAGCGUUCGCCGCAGGGCAAACGCGCCACUGCGGCAUCGGGGCUGUCCUCCG ---(-----------------((((((..................))))).)).(((((.((((((((((((((......))).)))))))....))))....))))) ( -28.37) >DroSim_CAF1 114 103 + 1 CAGAUUGCAUUGUC----CAGCUACUGGAAUACACC-GAUACUCACAAUGCGAUCGGAGCGCUCGCCGCAGGGCAAGCGGGCCACUGCGGACUCGGGACUGUCCUCCG ..((((((((((((----(((...))))).......-(.....).))))))))))((((..(((((((((((((......))).))))))...))))......)))). ( -42.30) >DroEre_CAF1 115 103 + 1 CAGAUUGCAUUGUC----CAGCUACUGGAAUACUCA-GAUACUCACAAUGCGAUCGGAGCGCUCGCCGCAGGGCAAGCGGGCCACUGCGGACUCGGGACUGUCCUCCG ..((((((((((((----(((...))))).......-(.....).))))))))))((((..(((((((((((((......))).))))))...))))......)))). ( -42.20) >DroWil_CAF1 36158 88 + 1 CAGGUUA-------------------GUAAUGGAGA-GUUACUCACAAUACGAUCAGAGCGUUCGCCGCAGGGCAAACGUGCUACUGCCGAUUCGGGACUAUCCUCGG .(((.((-------------------((..((((((-(...)))......((..(((.(((((.(((....))).)))))....))).)).))))..)))).)))... ( -23.00) >DroYak_CAF1 116 108 + 1 CAGAUUGCAUUGUAUCUGGAGCUACUGGAAUACACAAAAUACUCACAAUGCGAUCGGUGCGCUCGCCGCAGGGCAAGCGGGCCACUGCGGACUCGGGACUGUCCUCCG ..(((((((((((.....(((....((.......)).....))))))))))))))(((.((((.(((....))).)))).))).....((((........)))).... ( -39.00) >consensus CAGAUUGCAUUGUC____CAGCUACUGGAAUACACA_GAUACUCACAAUGCGAUCGGAGCGCUCGCCGCAGGGCAAGCGGGCCACUGCGGACUCGGGACUGUCCUCCG ..(((((((((((...............................)))))))))))((((..(((((((((((((......))).))))))...))))......)))). (-24.08 = -27.00 + 2.92)


| Location | 18,042,638 – 18,042,741 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.81 |
| Mean single sequence MFE | -41.38 |
| Consensus MFE | -29.42 |
| Energy contribution | -29.17 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18042638 103 + 20766785 GCGGAGGCGGUUGGGGCACCCAGUUGUAGGCCCGCCUGGAUAUGGGCACUCCAAAGCGAUCGUGCUGUGAGGCUUGUUGUCUGCAAA-GA--------------AAACAU-U-AAUUAUU ((((..((..((((((..((((..(.((((....)))).)..))))..)))))).))..))))(((....))).((((.(((....)-))--------------.)))).-.-....... ( -32.00) >DroPse_CAF1 337 114 + 1 GGGGCGGCGGCUGGGGCACCCAAUUGUAGGCCCGCCUAGAUAUGGGCACCCCGAAGCGGUCGUGCUGCGAGGCCUGUUGUCUGUGGAAGGAAAC---A---AGCACACGUCUCAGUUAGU ((.(((((.(((((((..((((..(.((((....)))).)..))))..))))..))).))))).))..(((((.(((.(..(((........))---)---..)))).)))))....... ( -47.90) >DroWil_CAF1 36246 99 + 1 GCGGCGGCGGUUGGGGUACCCAAUUGUAGGCCCGCCUCGAUAUAGGCACCCCAAAUCGAUCGUGCUGCGAUGCUUGUUGUCUGCAAUAGGAGAU------------CCUU-U-------- ((((((.((((((((((.((..((((.(((....)))))))...)).))))).)))))....))))))(((.(((((((....)))))))..))------------)...-.-------- ( -38.60) >DroMoj_CAF1 212 111 + 1 GUGGCGGCGGCUGGGGCACCCAGUUGUAGGCCCGCCUAGAGAUGGGCACCCCAAAGCGAUCAUGCUGCGACGCCUGUUGCCUGUGAA--AGUGGAAGUGGAAA---GAAU-U-AGUUA-- ..((((((((((((((..((((.((.((((....)))).)).))))..))))).(((......))).....))).))))))......--..............---....-.-.....-- ( -39.10) >DroAna_CAF1 219 112 + 1 GGGGCGGCGGCUGGGGCACCCAGUUGUAGGCCCGCCUCGAUAUGGGCACCCCGAAGCGAUCAUGCUGCGAGGCUUGUUGUCUGCAAA-AA-UAU---U---GGAACAGAUGUCAAUUAGU .((((.(((((((((...)))))))))..))))((((((...((((...)))).(((......))).))))))(((.(((((((((.-..-..)---)---)...)))))).)))..... ( -42.80) >DroPer_CAF1 338 114 + 1 GGGGCGGCGGCUGGGGCACCCAAUUGUAGGCCCGCCUAGAUAUGGGCACCCCGAAGCGGUCGUGCUGCGAGGCCUGUUGUCUGUGAAAGGAAAC---A---AGCACACGUCUCAGUUAGU ((.(((((.(((((((..((((..(.((((....)))).)..))))..))))..))).))))).))..(((((.(((.(..(((........))---)---..)))).)))))....... ( -47.90) >consensus GGGGCGGCGGCUGGGGCACCCAAUUGUAGGCCCGCCUAGAUAUGGGCACCCCAAAGCGAUCGUGCUGCGAGGCCUGUUGUCUGCAAA_GAAAAC_______AG_ACACAU_U_AGUUAGU ..((((((((((((((..((((..(.((((....)))).)..))))..))))).((((....)))).....))).))))))....................................... (-29.42 = -29.17 + -0.25)
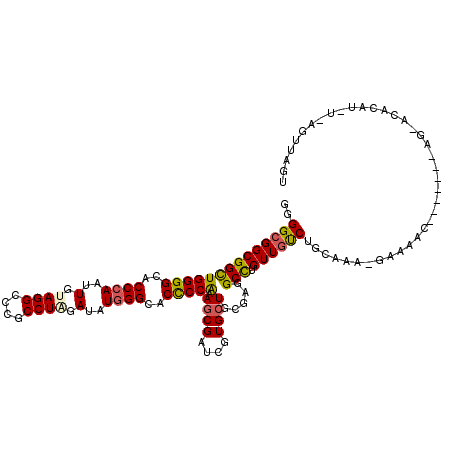
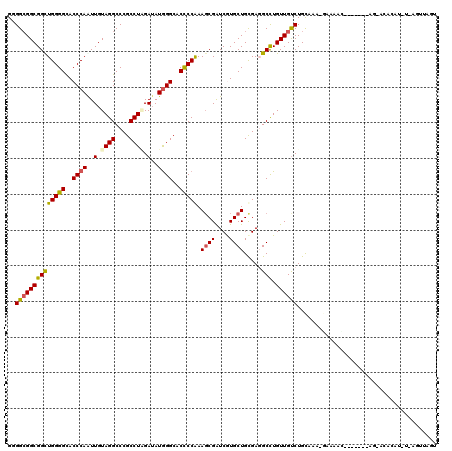
| Location | 18,042,638 – 18,042,741 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.81 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -25.06 |
| Energy contribution | -25.03 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.16 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
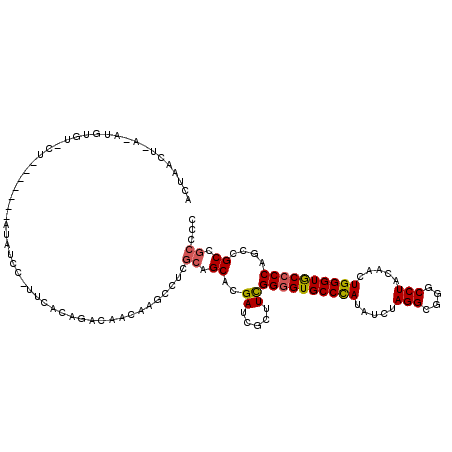
>2R_DroMel_CAF1 18042638 103 - 20766785 AAUAAUU-A-AUGUUU--------------UC-UUUGCAGACAACAAGCCUCACAGCACGAUCGCUUUGGAGUGCCCAUAUCCAGGCGGGCCUACAACUGGGUGCCCCAACCGCCUCCGC .......-.-.((((.--------------((-(....))).)))).(((((..(((......)))...))).))........(((((((((((....))))).......)))))).... ( -24.01) >DroPse_CAF1 337 114 - 1 ACUAACUGAGACGUGUGCU---U---GUUUCCUUCCACAGACAACAGGCCUCGCAGCACGACCGCUUCGGGGUGCCCAUAUCUAGGCGGGCCUACAAUUGGGUGCCCCAGCCGCCGCCCC .....(((.((.((.((.(---(---((((........)))))))).)).)).)))..((.(.(((..((((..((((....((((....))))....))))..))))))).).)).... ( -40.20) >DroWil_CAF1 36246 99 - 1 --------A-AAGG------------AUCUCCUAUUGCAGACAACAAGCAUCGCAGCACGAUCGAUUUGGGGUGCCUAUAUCGAGGCGGGCCUACAAUUGGGUACCCCAACCGCCGCCGC --------.-.(((------------....)))..(((.........)))..((.((.(....)..((((((((((((.....(((....))).....))))))))))))..)).))... ( -31.50) >DroMoj_CAF1 212 111 - 1 --UAACU-A-AUUC---UUUCCACUUCCACU--UUCACAGGCAACAGGCGUCGCAGCAUGAUCGCUUUGGGGUGCCCAUCUCUAGGCGGGCCUACAACUGGGUGCCCCAGCCGCCGCCAC --.....-.-....---..............--......(((....((((..(((((......))).(((((..((((....((((....))))....))))..)))))))))))))).. ( -39.40) >DroAna_CAF1 219 112 - 1 ACUAAUUGACAUCUGUUCC---A---AUA-UU-UUUGCAGACAACAAGCCUCGCAGCAUGAUCGCUUCGGGGUGCCCAUAUCGAGGCGGGCCUACAACUGGGUGCCCCAGCCGCCGCCCC .....(((...(((((...---.---...-..-...)))))...)))((...((.((..((.....))((((..((((.....(((....))).....))))..)))).)).)).))... ( -34.00) >DroPer_CAF1 338 114 - 1 ACUAACUGAGACGUGUGCU---U---GUUUCCUUUCACAGACAACAGGCCUCGCAGCACGACCGCUUCGGGGUGCCCAUAUCUAGGCGGGCCUACAAUUGGGUGCCCCAGCCGCCGCCCC .....(((.((.((.((.(---(---((((........)))))))).)).)).)))..((.(.(((..((((..((((....((((....))))....))))..))))))).).)).... ( -40.20) >consensus ACUAACU_A_AUGUGU_CU_______AUAUCC_UUCACAGACAACAAGCCUCGCAGCACGAUCGCUUCGGGGUGCCCAUAUCUAGGCGGGCCUACAACUGGGUGCCCCAGCCGCCGCCCC ....................................................((.((..((.....))((((((((((.....(((....))).....))))))))))....)).))... (-25.06 = -25.03 + -0.03)

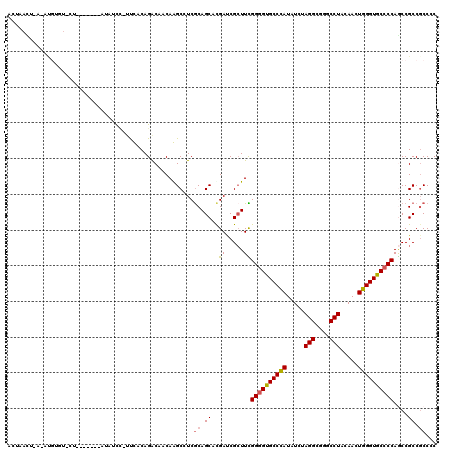
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:06 2006