| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,042,264 – 18,042,377 |
| Length | 113 |
| Max. P | 0.823395 |

| Location | 18,042,264 – 18,042,377 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -23.30 |
| Energy contribution | -23.17 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
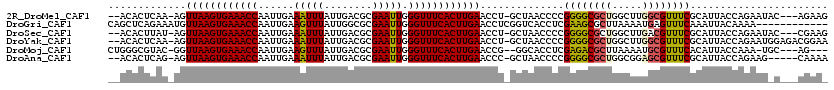
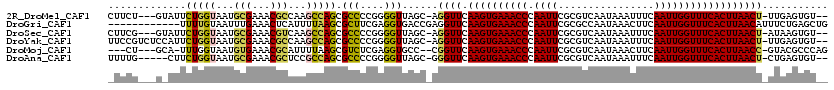
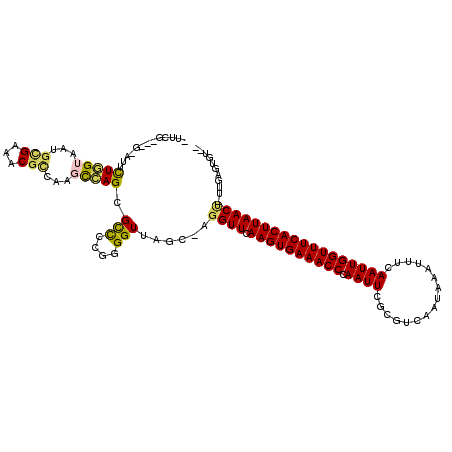
>2R_DroMel_CAF1 18042264 113 + 20766785 --ACACUCAA-AGUUAAGUGAAACCAAUUGAAAUUUAUUGACGCGAAUUGGGUUUCACUUGAACCU-GCUAACCCCGGGGCGCUGGCUUGGCGUUUCGCAUUACCAGAAUAC---AGAAG --....((..-..((((((((((((......(((((........))))).))))))))))))..((-(.(((...((..(((((.....)))))..))..))).))).....---.)).. ( -30.70) >DroGri_CAF1 75558 108 + 1 CAGCUCAGAAAUGUUAAGUGAAACCAAUUGAAGUUUAUUGGCGCGAAUUGGGUUUCACUUGAACCUCGGUCACCUCGAAGCGCUUAAAAUGAGUUUCAAAUUACAAAA------------ .((((((......((((((((((((((((...(((....)))...)))).))))))))))))...((((.....))))...........)))))).............------------ ( -24.60) >DroSec_CAF1 52828 113 + 1 --ACACUUAU-AGUUAAGUGAAACCAAUUGAAAUUUAUUGACGCGAAUUGGGUUUCACUUGAACCU-GCUAACCCCGGGGCGCUGGCUUGACGUUUCGCAUUACCAGAAUAC---CGAAG --........-..((((((((((((......(((((........))))).))))))))))))..((-(.(((...((..((((......).)))..))..))).))).....---..... ( -26.20) >DroYak_CAF1 53574 116 + 1 --ACACUCAA-AGUUAAGUGAAACCAAUUGAAAUUUAUUGACGCGAAUUGGGUUUCACUUGAACCU-GCUAACCCCGGGGCGCUGGCUUGGCGUUUCGCAUUACCAGAAUGGAGACGGAA --...(((..-..((((((((((((......(((((........))))).))))))))))))..((-(.(((...((..(((((.....)))))..))..))).)))....)))...... ( -33.80) >DroMoj_CAF1 70473 110 + 1 CUGGGCGUAC-GGUUAAGUGAAACCAAUUGAAGUUUAUUGACGCGAAUUGGGUUUCACUUGAACCG--GGCACCUCGAGACGCUUAAAAUGCGUUUCACAUUACCAAA-UGC---AG--- (((((.(..(-((((((((((((((((((...(((....)))...)))).))))))))))).))))--..).))..(((((((.......)))))))...........-..)---))--- ( -40.00) >DroAna_CAF1 47728 111 + 1 --ACACUCAG-AGUUAAGUGAAACCAAUUGAAAUUUAUUGACGCGAAUUGGGUUUCACUUGAACCC-GCUAACCCCGGGGCGCUGGCGGAGCGUUUCGCAUUACCAGAAG-----CAAAA --........-..((((((((((((......(((((........))))).)))))))))))).(((-(.......))))((.((((..(.(((...))).)..))))..)-----).... ( -31.90) >consensus __ACACUCAA_AGUUAAGUGAAACCAAUUGAAAUUUAUUGACGCGAAUUGGGUUUCACUUGAACCU_GCUAACCCCGGGGCGCUGGCUUGGCGUUUCGCAUUACCAGAAU_C___CGAA_ .............((((((((((((......(((((........))))).))))))))))))..............((((((((.....))))))))....................... (-23.30 = -23.17 + -0.14)
| Location | 18,042,264 – 18,042,377 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -19.33 |
| Energy contribution | -19.06 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
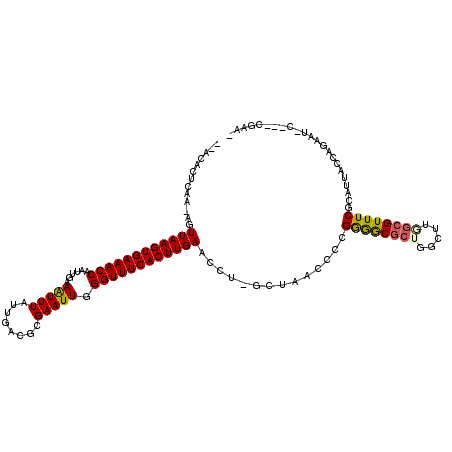
>2R_DroMel_CAF1 18042264 113 - 20766785 CUUCU---GUAUUCUGGUAAUGCGAAACGCCAAGCCAGCGCCCCGGGGUUAGC-AGGUUCAAGUGAAACCCAAUUCGCGUCAAUAAAUUUCAAUUGGUUUCACUUAACU-UUGAGUGU-- ...((---((...(((((...(((...)))...))))).(((....)))..))-))(((.((((((((((.((((................))))))))))))))))).-........-- ( -27.99) >DroGri_CAF1 75558 108 - 1 ------------UUUUGUAAUUUGAAACUCAUUUUAAGCGCUUCGAGGUGACCGAGGUUCAAGUGAAACCCAAUUCGCGCCAAUAAACUUCAAUUGGUUUCACUUAACAUUUCUGAGCUG ------------...............((((......(.((((((.(....))))))).)((((((((((.((((................))))))))))))))........))))... ( -24.99) >DroSec_CAF1 52828 113 - 1 CUUCG---GUAUUCUGGUAAUGCGAAACGUCAAGCCAGCGCCCCGGGGUUAGC-AGGUUCAAGUGAAACCCAAUUCGCGUCAAUAAAUUUCAAUUGGUUUCACUUAACU-AUAAGUGU-- (((((---(..(.(((((...(((...)))...))))).)..)))))).....-.((((.((((((((((.((((................))))))))))))))))))-........-- ( -28.99) >DroYak_CAF1 53574 116 - 1 UUCCGUCUCCAUUCUGGUAAUGCGAAACGCCAAGCCAGCGCCCCGGGGUUAGC-AGGUUCAAGUGAAACCCAAUUCGCGUCAAUAAAUUUCAAUUGGUUUCACUUAACU-UUGAGUGU-- ((((((..((.....))..))).)))(((((((((.(((.(....).))).))-.((((.((((((((((.((((................))))))))))))))))))-))).))))-- ( -29.19) >DroMoj_CAF1 70473 110 - 1 ---CU---GCA-UUUGGUAAUGUGAAACGCAUUUUAAGCGUCUCGAGGUGCC--CGGUUCAAGUGAAACCCAAUUCGCGUCAAUAAACUUCAAUUGGUUUCACUUAACC-GUACGCCCAG ---..---(((-((....)))))((.((((.......)))).))(.((((..--(((((.((((((((((.((((...((......))...))))))))))))))))))-)..))))).. ( -35.70) >DroAna_CAF1 47728 111 - 1 UUUUG-----CUUCUGGUAAUGCGAAACGCUCCGCCAGCGCCCCGGGGUUAGC-GGGUUCAAGUGAAACCCAAUUCGCGUCAAUAAAUUUCAAUUGGUUUCACUUAACU-CUGAGUGU-- .((((-----(..........)))))((((((.((.(((.(....).))).))-(((((.((((((((((.((((................))))))))))))))))))-).))))))-- ( -32.99) >consensus _UUCG___G_AUUCUGGUAAUGCGAAACGCCAAGCCAGCGCCCCGGGGUUAGC_AGGUUCAAGUGAAACCCAAUUCGCGUCAAUAAAUUUCAAUUGGUUUCACUUAACU_UUGAGUGU__ .............(((((...(((...)))...))))).(((....)))......((((.((((((((((.((((................))))))))))))))))))........... (-19.33 = -19.06 + -0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:03 2006