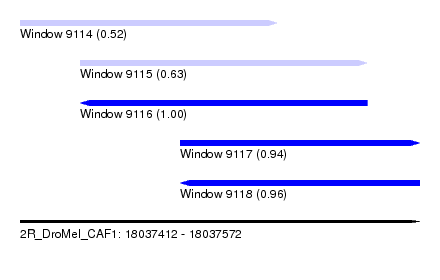
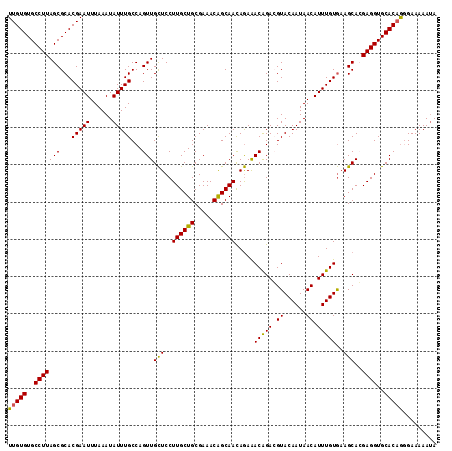
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,037,412 – 18,037,572 |
| Length | 160 |
| Max. P | 0.997205 |

| Location | 18,037,412 – 18,037,515 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.13 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -13.28 |
| Energy contribution | -13.76 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18037412 103 + 20766785 AACCUUUUUUAUAGC----------------AACACCCCUUAUUUUUCCUUGUGCACCUCCUGCUUCACAAAUGUUAUUGUACGUCUGUUUCUGUUGCUGUUUCGCAGCAAGGAGCAAC .............((----------------(.((...............)))))......(((((((((.((((......)))).))).....((((((.....)))))))))))).. ( -19.86) >DroSec_CAF1 48041 101 + 1 UAGCUUU-UUUUAGC----------------AGCACCCCUU-UUUUUCCCUGUGCACCUCGUGCUUCACAAAUGUUAUUGUACGUCUGUUUCUGUUGCCGUUUCGCAGCAAGGAGCAAC ..(((..-....)))----------------.((((.....-.........))))......(((((((((.((((......)))).)))...((((((......)))))).)))))).. ( -24.04) >DroSim_CAF1 47062 103 + 1 UAGCUUUUUUUUAGC----------------AGCACCCCUUAUUUUUCACUGUGCACCUCCUGCUUCACAAAUGUUAUUGUACGUCUGUUUCUGUUGCCGUUUCGCAGCAAGGAGCAAC ..(((.......)))----------------.((((...............))))......(((((((((.((((......)))).)))...((((((......)))))).)))))).. ( -23.36) >DroEre_CAF1 49772 119 + 1 CGCCGCUUUUUUAUCAGAAAACAUCCCAUGGAACACCCAUUAUUUUUCCCAGUGCACCUCGUGCUUCACAAAUGUUAUUGUACCUCUGUUUCUAUUGCUGUUUCGCAGCAAGGAGCAAC ....(((((......(((((.......((((.....)))).........(((.((((...))))...((((......))))....)))))))).((((((.....)))))))))))... ( -22.40) >DroYak_CAF1 48668 81 + 1 --------------------------------------UUUAUUUUUCGCUGUGCACCUCGUGCUUUACAAAUGUUAUUGUACCUCCGUCUUUGUUGCUGUUUCGCAGCAAGGAACAAC --------------------------------------............(((((((...))))..(((((......)))))......((((((((((......))))))))))))).. ( -18.20) >consensus UACCUUUUUUUUAGC________________AACACCCCUUAUUUUUCCCUGUGCACCUCGUGCUUCACAAAUGUUAUUGUACGUCUGUUUCUGUUGCUGUUUCGCAGCAAGGAGCAAC ...................................................(((((...(((.........)))....)))))....(((((((((((......))))).))))))... (-13.28 = -13.76 + 0.48)


| Location | 18,037,436 – 18,037,551 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -28.69 |
| Consensus MFE | -22.90 |
| Energy contribution | -23.58 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18037436 115 + 20766785 UAUUUUUCCUUGUGCACCUCCUGCUUCACAAAUGUUAUUGUACGUCUGUUUCUGUUGCUGUUUCGCAGCAAGGAGCAACUGGCAAAUAUUUAAAUUCGUGCGCUAAGGCACACAA .......((((((((((..........((((......))))..(((((((((((((((......))))).)))))))...)))..............)))))).))))....... ( -29.10) >DroSec_CAF1 48064 114 + 1 U-UUUUUCCCUGUGCACCUCGUGCUUCACAAAUGUUAUUGUACGUCUGUUUCUGUUGCCGUUUCGCAGCAAGGAGCAACUGGCAAAUAUUUAAAUUCGUGCGCUAAGGCACACAA .-......(((((((((...((((...((....))....))))(((((((((((((((......))))).)))))))...)))..............))))))..)))....... ( -29.10) >DroSim_CAF1 47086 115 + 1 UAUUUUUCACUGUGCACCUCCUGCUUCACAAAUGUUAUUGUACGUCUGUUUCUGUUGCCGUUUCGCAGCAAGGAGCAACUGGCAAAUAUUUAAAUUCGUGCGCUAAGGCACACAA ............(((......(((((((((.((((......)))).)))...((((((......)))))).))))))....))).............(((.((....)).))).. ( -26.40) >DroEre_CAF1 49812 115 + 1 UAUUUUUCCCAGUGCACCUCGUGCUUCACAAAUGUUAUUGUACCUCUGUUUCUAUUGCUGUUUCGCAGCAAGGAGCAACUGGCAAAUAUUUAAAUUCGUGCGCUAAGGCACACAA ....(((.(((((((((...))))...((((......)))).....(((((((..(((((.....))))))))))))))))).)))...........(((.((....)).))).. ( -29.30) >DroYak_CAF1 48670 115 + 1 UAUUUUUCGCUGUGCACCUCGUGCUUUACAAAUGUUAUUGUACCUCCGUCUUUGUUGCUGUUUCGCAGCAAGGAACAACUGGCAAAUAUUUAAAUUCGUGCGCUAAGGCACACAA ........(.(((((....((..(.....(((((((..(((.......((((((((((......)))))))))).......))))))))))......)..)).....)))))).. ( -29.54) >consensus UAUUUUUCCCUGUGCACCUCGUGCUUCACAAAUGUUAUUGUACGUCUGUUUCUGUUGCUGUUUCGCAGCAAGGAGCAACUGGCAAAUAUUUAAAUUCGUGCGCUAAGGCACACAA ...........((((.....((((.....(((((((..(((.....((((((((((((......))))).)))))))....))))))))))........))))....)))).... (-22.90 = -23.58 + 0.68)

| Location | 18,037,436 – 18,037,551 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -28.20 |
| Energy contribution | -27.68 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18037436 115 - 20766785 UUGUGUGCCUUAGCGCACGAAUUUAAAUAUUUGCCAGUUGCUCCUUGCUGCGAAACAGCAACAGAAACAGACGUACAAUAACAUUUGUGAAGCAGGAGGUGCACAAGGAAAAAUA (((((..((((.((.(((((((..............(((.((..((((((.....)))))).)).)))....((......)))))))))..))..))))..)))))......... ( -30.20) >DroSec_CAF1 48064 114 - 1 UUGUGUGCCUUAGCGCACGAAUUUAAAUAUUUGCCAGUUGCUCCUUGCUGCGAAACGGCAACAGAAACAGACGUACAAUAACAUUUGUGAAGCACGAGGUGCACAGGGAAAAA-A ((((((((....)))))))).........(((.((.((.((.(((((.(((.....(....)....(((((.((......)).)))))...)))))))).)))).)).)))..-. ( -33.30) >DroSim_CAF1 47086 115 - 1 UUGUGUGCCUUAGCGCACGAAUUUAAAUAUUUGCCAGUUGCUCCUUGCUGCGAAACGGCAACAGAAACAGACGUACAAUAACAUUUGUGAAGCAGGAGGUGCACAGUGAAAAAUA (((((..((((.((.(((((((........(((((..((((........))))...)))))...........((......)))))))))..))..))))..)))))......... ( -32.10) >DroEre_CAF1 49812 115 - 1 UUGUGUGCCUUAGCGCACGAAUUUAAAUAUUUGCCAGUUGCUCCUUGCUGCGAAACAGCAAUAGAAACAGAGGUACAAUAACAUUUGUGAAGCACGAGGUGCACUGGGAAAAAUA ((((((((....)))))))).........(((.(((((.((.((((((((.....))))..........(...(((((......)))))...)..)))).))))))).))).... ( -35.80) >DroYak_CAF1 48670 115 - 1 UUGUGUGCCUUAGCGCACGAAUUUAAAUAUUUGCCAGUUGUUCCUUGCUGCGAAACAGCAACAAAGACGGAGGUACAAUAACAUUUGUAAAGCACGAGGUGCACAGCGAAAAAUA (((((..((((.((..((((((.....((((((((.(((.....((((((.....))))))....)))...)))).))))..))))))...))..))))..)))))......... ( -30.90) >consensus UUGUGUGCCUUAGCGCACGAAUUUAAAUAUUUGCCAGUUGCUCCUUGCUGCGAAACAGCAACAGAAACAGACGUACAAUAACAUUUGUGAAGCACGAGGUGCACAGGGAAAAAUA (((((..(((((((...(((((......)))))...)))(((..((((((.....)))))).....(((((.((......)).)))))..)))..))))..)))))......... (-28.20 = -27.68 + -0.52)


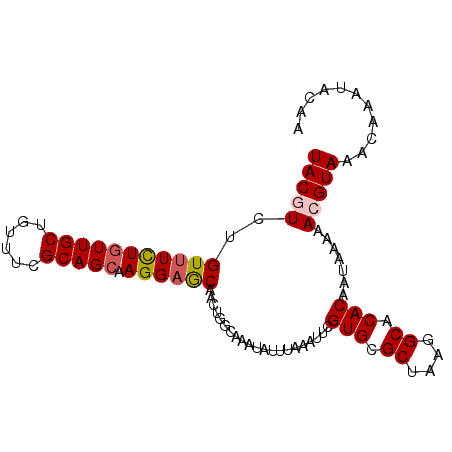
| Location | 18,037,476 – 18,037,572 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.76 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18037476 96 + 20766785 UACGUCUGUUUCUGUUGCUGUUUCGCAGCAAGGAGCAACUGGCAAAUAUUUAAAUUCGUGCGCUAAGGCACACAAUAAAAACGUAAACAAAUACAA (((((.((((((((((((......))))).)))))))....................(((.((....)).))).......)))))........... ( -23.40) >DroSec_CAF1 48103 96 + 1 UACGUCUGUUUCUGUUGCCGUUUCGCAGCAAGGAGCAACUGGCAAAUAUUUAAAUUCGUGCGCUAAGGCACACAAUAAAAACGUAAACAAAUACAA (((((.((((((((((((......))))).)))))))....................(((.((....)).))).......)))))........... ( -22.90) >DroSim_CAF1 47126 96 + 1 UACGUCUGUUUCUGUUGCCGUUUCGCAGCAAGGAGCAACUGGCAAAUAUUUAAAUUCGUGCGCUAAGGCACACAAUAAAAACGUAAACAAAUACAA (((((.((((((((((((......))))).)))))))....................(((.((....)).))).......)))))........... ( -22.90) >DroEre_CAF1 49852 96 + 1 UACCUCUGUUUCUAUUGCUGUUUCGCAGCAAGGAGCAACUGGCAAAUAUUUAAAUUCGUGCGCUAAGGCACACAAUAAAAACGUAAACAAAUACAA ..((..(((((((..(((((.....))))))))))))...))...............(((.((....)).))).........(((......))).. ( -21.20) >DroYak_CAF1 48710 96 + 1 UACCUCCGUCUUUGUUGCUGUUUCGCAGCAAGGAACAACUGGCAAAUAUUUAAAUUCGUGCGCUAAGGCACACAAUAAAAACGUAAACAAAUACAA ..((....((((((((((......))))))))))......))...............(((.((....)).))).........(((......))).. ( -21.20) >consensus UACGUCUGUUUCUGUUGCUGUUUCGCAGCAAGGAGCAACUGGCAAAUAUUUAAAUUCGUGCGCUAAGGCACACAAUAAAAACGUAAACAAAUACAA (((((..(((((((((((......))))).)))))).....................(((.((....)).))).......)))))........... (-18.28 = -18.76 + 0.48)


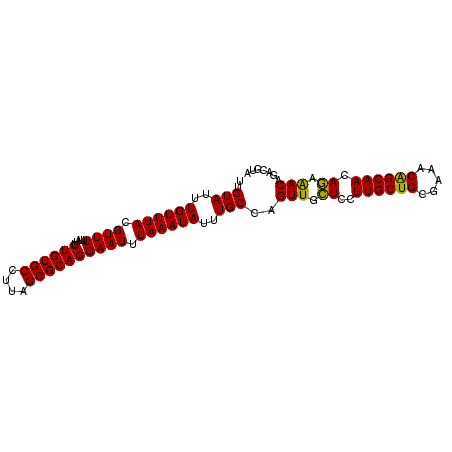
| Location | 18,037,476 – 18,037,572 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -21.92 |
| Energy contribution | -21.20 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18037476 96 - 20766785 UUGUAUUUGUUUACGUUUUUAUUGUGUGCCUUAGCGCACGAAUUUAAAUAUUUGCCAGUUGCUCCUUGCUGCGAAACAGCAACAGAAACAGACGUA ...........(((((((...((((((((....))))))))................(((.((..((((((.....)))))).)).)))))))))) ( -23.40) >DroSec_CAF1 48103 96 - 1 UUGUAUUUGUUUACGUUUUUAUUGUGUGCCUUAGCGCACGAAUUUAAAUAUUUGCCAGUUGCUCCUUGCUGCGAAACGGCAACAGAAACAGACGUA ((((.((((((..((((((..((((((((....))))))))............((.(((........)))))))))))..)))))).))))..... ( -26.00) >DroSim_CAF1 47126 96 - 1 UUGUAUUUGUUUACGUUUUUAUUGUGUGCCUUAGCGCACGAAUUUAAAUAUUUGCCAGUUGCUCCUUGCUGCGAAACGGCAACAGAAACAGACGUA ((((.((((((..((((((..((((((((....))))))))............((.(((........)))))))))))..)))))).))))..... ( -26.00) >DroEre_CAF1 49852 96 - 1 UUGUAUUUGUUUACGUUUUUAUUGUGUGCCUUAGCGCACGAAUUUAAAUAUUUGCCAGUUGCUCCUUGCUGCGAAACAGCAAUAGAAACAGAGGUA ..(((......)))(((((((((((((((......))))...........(((((.(((........))))))))...)))))))))))....... ( -23.20) >DroYak_CAF1 48710 96 - 1 UUGUAUUUGUUUACGUUUUUAUUGUGUGCCUUAGCGCACGAAUUUAAAUAUUUGCCAGUUGUUCCUUGCUGCGAAACAGCAACAAAGACGGAGGUA .............(((((((.((((((((....))))))))........................((((((.....)))))).)))))))...... ( -22.50) >consensus UUGUAUUUGUUUACGUUUUUAUUGUGUGCCUUAGCGCACGAAUUUAAAUAUUUGCCAGUUGCUCCUUGCUGCGAAACAGCAACAGAAACAGACGUA ..(((..((((((.((((.....((((((....)))))))))).))))))..)))..(((.((..((((((.....)))))).)).)))....... (-21.92 = -21.20 + -0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:00 2006