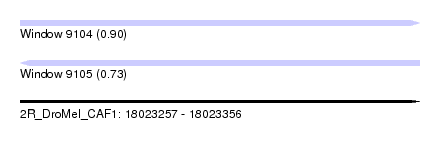
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,023,257 – 18,023,356 |
| Length | 99 |
| Max. P | 0.896198 |

| Location | 18,023,257 – 18,023,356 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 91.32 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -25.76 |
| Energy contribution | -25.80 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18023257 99 + 20766785 -----UUUGAGUGCCGGAACAAAGGCAGCCUUCAUGAUAUCCUCGGAUGAGCAUUUGGCUGGUAUGCAGCUUCAUUCUCAUGGGUUGCUUCUGGU----UCCCGUCCU -----....((..(.(((((..((((((((..(((((.......((((((......(((((.....))))))))))))))))))))))))...))----))).)..)) ( -33.31) >DroSec_CAF1 37243 99 + 1 -----UUUGAGUGCCGGAACAAAGGCAGCCUUCAUGAUAUCCUCGGAUGAGCAUUUGGCUGCUAUGCAGCUUCAUUCUCAUGCGCUGCUUCUGGU----UCCCCUCCU -----...(((.(((((((....(((((((...(((.((((....))))..)))..)))))))..(((((..(((....))).))))))))))))----....))).. ( -35.00) >DroSim_CAF1 36270 103 + 1 -----UUUGAGUGCCGGAACAAAGGCAGCCUUCAUGAUAUCCUCGGAUGAGCAUUUGGCUGGUAUGCAGCUUCAUUCUCAUGCGCUGCUUCUGGUCUUCUUCCUUCCU -----...(((.((((((.....((((((......((.....))(.(((((.((..(((((.....)))))..)).))))).))))))))))))).)))......... ( -31.90) >DroEre_CAF1 38787 98 + 1 -----UUUGAGUGCCGGAACAAAGACAGCCUUCAUGAUAUCCUCGGAUGAGCAUUUGGCUGGUAUGCAGCUUCAUUCUCAUGGGCUGCUUCUGGU----UCCCCUCU- -----...(((.(((((((......(((((...(((.((((....))))..)))..)))))....((((((.(((....))))))))))))))))----....))).- ( -33.20) >DroYak_CAF1 36670 103 + 1 GUGCGUUUGAGUGGCGGAACAAAGACAGCCUUCAUGAUAUCCUCGGAUGAGCAUUUGGCUGGUAUGCAGCUUCAUUCUCAUGCGCUGCUUCUGGU----UCCGCUGC- ..........(..(((((((..((((((((...(((.((((....))))..)))..)))))....(((((..(((....))).))))).))).))----)))))..)- ( -37.50) >consensus _____UUUGAGUGCCGGAACAAAGGCAGCCUUCAUGAUAUCCUCGGAUGAGCAUUUGGCUGGUAUGCAGCUUCAUUCUCAUGCGCUGCUUCUGGU____UCCCCUCCU ............(((((((......(((((...(((.((((....))))..)))..)))))....(((((.............))))))))))))............. (-25.76 = -25.80 + 0.04)

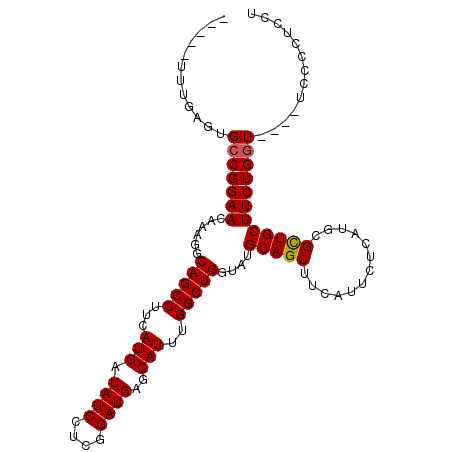
| Location | 18,023,257 – 18,023,356 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 91.32 |
| Mean single sequence MFE | -31.86 |
| Consensus MFE | -22.90 |
| Energy contribution | -22.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18023257 99 - 20766785 AGGACGGGA----ACCAGAAGCAACCCAUGAGAAUGAAGCUGCAUACCAGCCAAAUGCUCAUCCGAGGAUAUCAUGAAGGCUGCCUUUGUUCCGGCACUCAAA----- ((..(.(((----((.(((.(((.(((((((.......((((.....))))...((.(((....))).)).)))))..)).))).)))))))).)..))....----- ( -27.10) >DroSec_CAF1 37243 99 - 1 AGGAGGGGA----ACCAGAAGCAGCGCAUGAGAAUGAAGCUGCAUAGCAGCCAAAUGCUCAUCCGAGGAUAUCAUGAAGGCUGCCUUUGUUCCGGCACUCAAA----- ..(((.(((----((.(((.(((((.(((((.......(((((...)))))...((.(((....))).)).)))))...))))).))))))))....)))...----- ( -34.30) >DroSim_CAF1 36270 103 - 1 AGGAAGGAAGAAGACCAGAAGCAGCGCAUGAGAAUGAAGCUGCAUACCAGCCAAAUGCUCAUCCGAGGAUAUCAUGAAGGCUGCCUUUGUUCCGGCACUCAAA----- .((((.((((..........(((((.(((....)))..)))))....(((((..(((...(((....)))..)))...))))).)))).))))..........----- ( -30.90) >DroEre_CAF1 38787 98 - 1 -AGAGGGGA----ACCAGAAGCAGCCCAUGAGAAUGAAGCUGCAUACCAGCCAAAUGCUCAUCCGAGGAUAUCAUGAAGGCUGUCUUUGUUCCGGCACUCAAA----- -.(((.(((----((.(((..((((((((((.......((((.....))))...((.(((....))).)).)))))..)))))..))))))))....)))...----- ( -32.90) >DroYak_CAF1 36670 103 - 1 -GCAGCGGA----ACCAGAAGCAGCGCAUGAGAAUGAAGCUGCAUACCAGCCAAAUGCUCAUCCGAGGAUAUCAUGAAGGCUGUCUUUGUUCCGCCACUCAAACGCAC -...(((((----((.(((.(((((.(((....)))..)))))....(((((..(((...(((....)))..)))...))))))))..)))))))............. ( -34.10) >consensus AGGAGGGGA____ACCAGAAGCAGCGCAUGAGAAUGAAGCUGCAUACCAGCCAAAUGCUCAUCCGAGGAUAUCAUGAAGGCUGCCUUUGUUCCGGCACUCAAA_____ .....((((......((((.(((((.(((((.......((((.....))))...((.(((....))).)).)))))...))))).))))))))............... (-22.90 = -22.90 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:47 2006