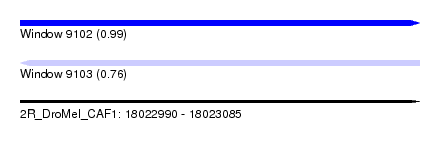
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,022,990 – 18,023,085 |
| Length | 95 |
| Max. P | 0.992283 |

| Location | 18,022,990 – 18,023,085 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 92.84 |
| Mean single sequence MFE | -26.66 |
| Consensus MFE | -25.04 |
| Energy contribution | -24.96 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18022990 95 + 20766785 UUAACCACGAACAUGUAUAUGCCAUGCUCAAUUAGGCGAUCCUUUUCAAAACGCCGUGUCUGAAGAUCGUUUUGAGCGGGUUGCCAACCCGUAAU ......((((.((((.......)))).)).....((((((((..(((((((((....(((....)))))))))))).)))))))).....))... ( -26.50) >DroSec_CAF1 36976 95 + 1 UCAACCGUGAACAUGUAUAUGUCAUGCUCAAUUAGGCGAUCCCUAUCAAAACGCCGGGUCUGAAGAUCGUUUUGAGCGGGUUGCCAAUCAGUAAU .......(((.((((.......)))).)))....((((((((...((((((((....(((....)))))))))))..)))))))).......... ( -28.00) >DroSim_CAF1 36003 95 + 1 UCAACCAUGAACAUGUAUAUGUCAUGCUCAAUUAGGCGAUCCUUUUCAGAACGCCGGGUCUGAAGAUCGUUUUGAGCGGGUUGCCAACCAGUAAU .(((((((((.(((....)))))))((((((...(((((((...((((((.(....).))))))))))))))))))).)))))............ ( -29.20) >DroEre_CAF1 38521 95 + 1 UUAACCAUAAACAUGUAUAUGUCAUGCUCAAUUAGGCGAUCCUUUUCAAAACGCCGGGUCUGAAGAUCGUUUUGAGCGGGUUGCCACCCCGAAAU ...........((((.......))))........((((((((..(((((((((....(((....)))))))))))).)))))))).......... ( -24.50) >DroYak_CAF1 36404 95 + 1 UUAACCAUGAACAUGUAUAUGUCAUGCUCAAUUAGGCGAUCCUUUUUGAAACGCCGGGUCUGAAGAUCGUUUUGAGCGGGUUGCCACCCCGAAAU .......(((.((((.......)))).)))....((((((((..((..(((((....(((....))))))))..)).)))))))).......... ( -25.10) >consensus UUAACCAUGAACAUGUAUAUGUCAUGCUCAAUUAGGCGAUCCUUUUCAAAACGCCGGGUCUGAAGAUCGUUUUGAGCGGGUUGCCAACCCGUAAU .......(((.((((.......)))).)))....((((((((...((((((((....(((....)))))))))))..)))))))).......... (-25.04 = -24.96 + -0.08)

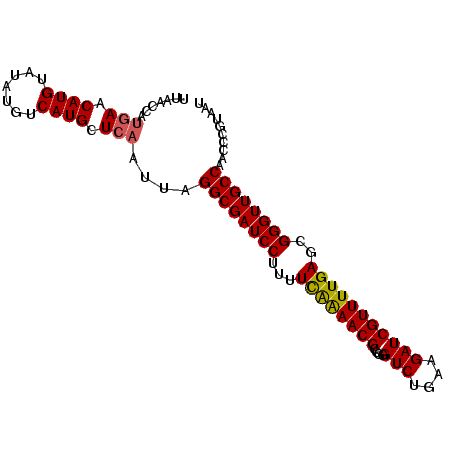
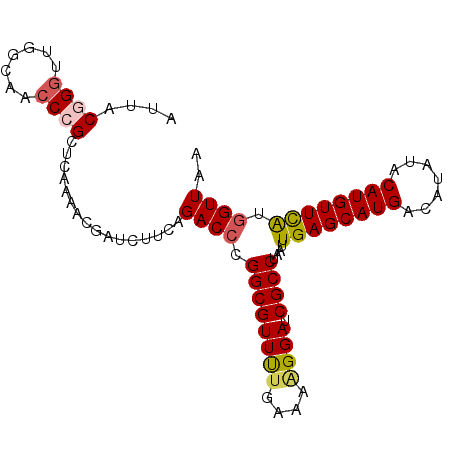
| Location | 18,022,990 – 18,023,085 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 92.84 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -20.98 |
| Energy contribution | -21.38 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18022990 95 - 20766785 AUUACGGGUUGGCAACCCGCUCAAAACGAUCUUCAGACACGGCGUUUUGAAAAGGAUCGCCUAAUUGAGCAUGGCAUAUACAUGUUCGUGGUUAA ....((..(((((.....)).)))..)).........(((((((((((....)))).)))).....(((((((.......))))))))))..... ( -28.10) >DroSec_CAF1 36976 95 - 1 AUUACUGAUUGGCAACCCGCUCAAAACGAUCUUCAGACCCGGCGUUUUGAUAGGGAUCGCCUAAUUGAGCAUGACAUAUACAUGUUCACGGUUGA ...((((...(((..(((..((((((((.((.........))))))))))..)))...)))....((((((((.......))))))))))))... ( -28.30) >DroSim_CAF1 36003 95 - 1 AUUACUGGUUGGCAACCCGCUCAAAACGAUCUUCAGACCCGGCGUUCUGAAAAGGAUCGCCUAAUUGAGCAUGACAUAUACAUGUUCAUGGUUGA ............(((((.((((((.....((....))...((((((((....)))).))))...))))))(((((((....))).))))))))). ( -27.30) >DroEre_CAF1 38521 95 - 1 AUUUCGGGGUGGCAACCCGCUCAAAACGAUCUUCAGACCCGGCGUUUUGAAAAGGAUCGCCUAAUUGAGCAUGACAUAUACAUGUUUAUGGUUAA ......((((((....)))))).............((((.((((((((....)))).))))....((((((((.......)))))))).)))).. ( -26.00) >DroYak_CAF1 36404 95 - 1 AUUUCGGGGUGGCAACCCGCUCAAAACGAUCUUCAGACCCGGCGUUUCAAAAAGGAUCGCCUAAUUGAGCAUGACAUAUACAUGUUCAUGGUUAA ......((((((....)))))).............((((.((((.(((.....))).))))....((((((((.......)))))))).)))).. ( -27.80) >consensus AUUACGGGUUGGCAACCCGCUCAAAACGAUCUUCAGACCCGGCGUUUUGAAAAGGAUCGCCUAAUUGAGCAUGACAUAUACAUGUUCAUGGUUAA ....((((.......))))................((((.((((((((....)))).))))....((((((((.......)))))))).)))).. (-20.98 = -21.38 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:45 2006