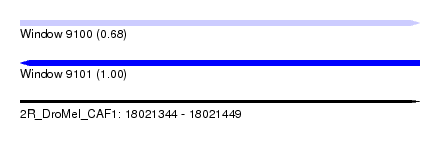
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,021,344 – 18,021,449 |
| Length | 105 |
| Max. P | 0.997718 |

| Location | 18,021,344 – 18,021,449 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -18.79 |
| Energy contribution | -18.99 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18021344 105 + 20766785 CACACGAUUCUCACGCCUCAAGUGGCGGCCAUAAUUACUGCAAUC----UCGUAAUCGUAAUCACAACUGUAAUUGUAAUUCAAUUCGCUCGGCCGCCUCCG-CACUCAA- ..............((.....(.(((((((...((((((((....----..)))...))))).......(((((((.....))))).))..))))))).).)-)......- ( -21.30) >DroSec_CAF1 35230 105 + 1 CACAUGAUUCUCACGCCUUAAGUGGCGGCCAUAAUUACUGCAAUC----UCGUAAUCGUAAUCACAACUGUAAUUGUAAUUCAAUUCGCUCGGCCGCCUCCG-CACUCAA- ..............((.....(.(((((((...((((((((....----..)))...))))).......(((((((.....))))).))..))))))).).)-)......- ( -21.30) >DroSim_CAF1 34260 105 + 1 CACACGAUUCUCACGCCUCAAGUGGCGGCCAUAAUUACUGCAAUC----UCGUAAUCGUAAUCACAACUGUAAUUGUAAUUCAAUUCGCUCGGCCGCCUCCG-CACUCAA- ..............((.....(.(((((((...((((((((....----..)))...))))).......(((((((.....))))).))..))))))).).)-)......- ( -21.30) >DroEre_CAF1 36755 105 + 1 CACACGAUUCUCACGCCUCAAGUGGCGGCCAUAAUUACUGCAAUC----UCGUAAUCGUAAUCACAACUGUAAUUGUAAUUCAAUUCGCUCGGCCGCCUCCG-CACUCAA- ..............((.....(.(((((((...((((((((....----..)))...))))).......(((((((.....))))).))..))))))).).)-)......- ( -21.30) >DroYak_CAF1 34710 105 + 1 CACACGAUUCUCACGCCUCAAGUGGCGGCCAUAAUUACUGCAAUC----UCGUAAUCGUAAUCACAACUGUAAUUGUAAUUCAAUUCGCUCGGCCGCCUCCG-CACUUAA- ..............((.....(.(((((((...((((((((....----..)))...))))).......(((((((.....))))).))..))))))).).)-)......- ( -21.30) >DroAna_CAF1 29815 111 + 1 CACACAAUUCUCACGCCUCAAGUGGCGGCCAUAAUUACUGCAAUCGUAAUCGUUGUCGUAAUCGCAACUGUAAUUGUAAUUCAAUUCGCUUGGCCACCUCCUCCACUCACU ..............(((......)))(((((..(((((.((((.((....)))))).))))).......(((((((.....))))).)).)))))................ ( -23.40) >consensus CACACGAUUCUCACGCCUCAAGUGGCGGCCAUAAUUACUGCAAUC____UCGUAAUCGUAAUCACAACUGUAAUUGUAAUUCAAUUCGCUCGGCCGCCUCCG_CACUCAA_ .....................(.((((((((..(((((.............)))))..)..........(((((((.....))))).))..))))))).)........... (-18.79 = -18.99 + 0.20)
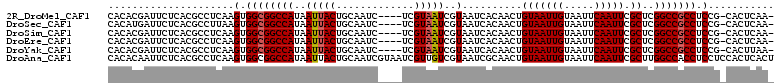
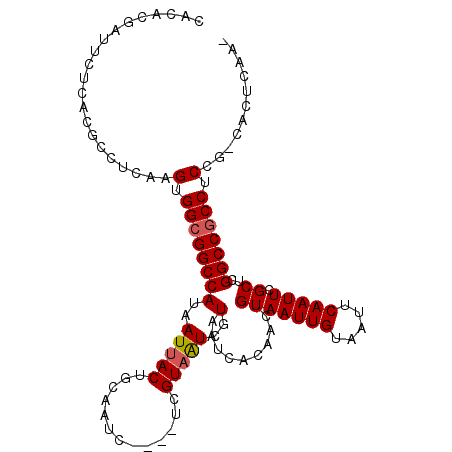
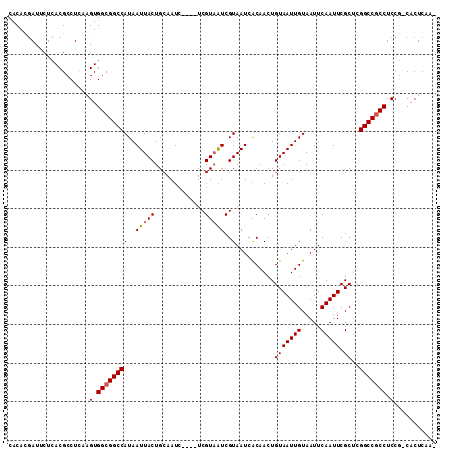
| Location | 18,021,344 – 18,021,449 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -31.04 |
| Energy contribution | -31.18 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18021344 105 - 20766785 -UUGAGUG-CGGAGGCGGCCGAGCGAAUUGAAUUACAAUUACAGUUGUGAUUACGAUUACGA----GAUUGCAGUAAUUAUGGCCGCCACUUGAGGCGUGAGAAUCGUGUG -....(..-(...((((((((..((((((((((((((((....))))))))).))))).)).----(((((...))))).))))))))....((..(....)..)))..). ( -34.10) >DroSec_CAF1 35230 105 - 1 -UUGAGUG-CGGAGGCGGCCGAGCGAAUUGAAUUACAAUUACAGUUGUGAUUACGAUUACGA----GAUUGCAGUAAUUAUGGCCGCCACUUAAGGCGUGAGAAUCAUGUG -((((((.-....((((((((..((((((((((((((((....))))))))).))))).)).----(((((...))))).)))))))))))))).(((((.....))))). ( -34.40) >DroSim_CAF1 34260 105 - 1 -UUGAGUG-CGGAGGCGGCCGAGCGAAUUGAAUUACAAUUACAGUUGUGAUUACGAUUACGA----GAUUGCAGUAAUUAUGGCCGCCACUUGAGGCGUGAGAAUCGUGUG -....(..-(...((((((((..((((((((((((((((....))))))))).))))).)).----(((((...))))).))))))))....((..(....)..)))..). ( -34.10) >DroEre_CAF1 36755 105 - 1 -UUGAGUG-CGGAGGCGGCCGAGCGAAUUGAAUUACAAUUACAGUUGUGAUUACGAUUACGA----GAUUGCAGUAAUUAUGGCCGCCACUUGAGGCGUGAGAAUCGUGUG -....(..-(...((((((((..((((((((((((((((....))))))))).))))).)).----(((((...))))).))))))))....((..(....)..)))..). ( -34.10) >DroYak_CAF1 34710 105 - 1 -UUAAGUG-CGGAGGCGGCCGAGCGAAUUGAAUUACAAUUACAGUUGUGAUUACGAUUACGA----GAUUGCAGUAAUUAUGGCCGCCACUUGAGGCGUGAGAAUCGUGUG -....(..-(...((((((((..((((((((((((((((....))))))))).))))).)).----(((((...))))).))))))))....((..(....)..)))..). ( -34.10) >DroAna_CAF1 29815 111 - 1 AGUGAGUGGAGGAGGUGGCCAAGCGAAUUGAAUUACAAUUACAGUUGCGAUUACGACAACGAUUACGAUUGCAGUAAUUAUGGCCGCCACUUGAGGCGUGAGAAUUGUGUG ........(((..((((((((.((.((((((((....))).)))))))((((((..(((((....)).)))..)))))).)))))))).)))((..(....)..))..... ( -31.20) >consensus _UUGAGUG_CGGAGGCGGCCGAGCGAAUUGAAUUACAAUUACAGUUGUGAUUACGAUUACGA____GAUUGCAGUAAUUAUGGCCGCCACUUGAGGCGUGAGAAUCGUGUG .............((((((((..((((((((((((((((....))))))))).))))).)).....(((((...))))).))))))))((.(((..(....)..))).)). (-31.04 = -31.18 + 0.14)

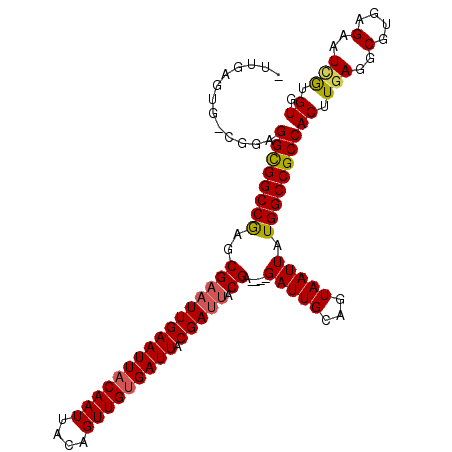
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:44 2006