| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,005,177 – 18,005,287 |
| Length | 110 |
| Max. P | 0.712326 |

| Location | 18,005,177 – 18,005,287 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.58 |
| Mean single sequence MFE | -45.25 |
| Consensus MFE | -25.49 |
| Energy contribution | -26.77 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
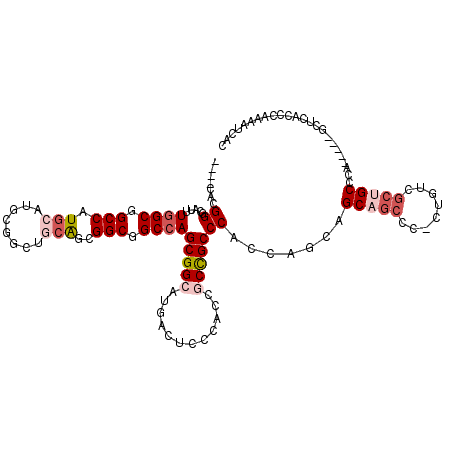
>2R_DroMel_CAF1 18005177 110 - 20766785 ----CACGGGCAUUUGGCGGCCAUGCAUGCGGCUGCAGCGGCGGCAAGCGGAAUGACUCCGACCGCUGCCCAUCAACAGCAGCCC-CUGUCGCUGCCCA-----GCUCACCCAAAAUCAC ----...((((....((((((...(((.(.((((((.(((((((....((((.....)))).))))))).........)))))))-.))).))))))..-----))))............ ( -44.80) >DroVir_CAF1 25623 113 - 1 ----CAUGGCCAUCUGGCGGCCAUACAUGCAG---CGGCGGCUGCCAGCGGCUUGACACCCGUCGCCGCCCAUCAGCAGCCCAUAAGCGCCGCAGCUCAUCCGACGUCGCCCAAGAUUAC ----.......((((((((((.......((.(---((((((((((..(((((..(((....))))))))......)))))).......))))).)).........))))))..))))... ( -42.40) >DroEre_CAF1 20614 110 - 1 ----CACGGGCACUUGGCGGCCAUGCAUGCGGCUGCAGCGGCGGCCAGCGGAAUGACUCCAACGGCUGCCCACCAGCAGCAGCCC-CUGUCGCUGCCCA-----GCUCCCCCAAAAUCAC ----...(((..((.((((((...(((.(.((((((.(((((((((...(((.....)))...))))))).....)).)))))))-.))).)))))).)-----)..))).......... ( -49.00) >DroYak_CAF1 17928 110 - 1 ----CACGGGCAUUUGGCGGCCAUGCAUGCGGCUGCAGCGGCGGCCAGCGGCAUGACUCCCACAGCUGCCCACCAGCAGCAGCCC-CUGUCGCUGCCCA-----GCUCACCCAAAAUCAC ----((.((((...(((.((.(((((.(((((((((....)))))).)))))))).)).)))..(((((......))))).))))-.))..(((....)-----)).............. ( -46.00) >DroMoj_CAF1 24621 113 - 1 ----CAUGGACACCUGGCCGCCAUGCACGCAG---CAGCGGCUGCCAGCGGCCUCACUCCUGUCGCCGCCCAUCAGCAGCCGCUUAGUGGCGCUGCGCAUCCGACGUCGCCGAAGAUUAC ----..((((..(.((((.(((((((.....)---)(((((((((..(((((..((....))..)))))......)))))))))..))))))))).)..))))..(((......)))... ( -43.50) >DroAna_CAF1 15116 114 - 1 GAGUCACGGCCAUUUGGCGGCCAUGCAUGCGGCCGCAGCGGCUGCCAGCGGCCUCACUCCGACGGCCGCCCACCAACAGCAGCCC-AUUUCGCUGCCCA-----GCUCUCCGAAGAUCAC ((((...((.....((((((((..((.(((....)))))))))))))((((((((.....)).))))))...))....(((((..-.....)))))...-----))))............ ( -45.80) >consensus ____CACGGGCAUUUGGCGGCCAUGCAUGCGGCUGCAGCGGCGGCCAGCGGCAUGACUCCCACCGCCGCCCACCAGCAGCAGCCC_CUGUCGCUGCCCA_____GCUCACCCAAAAUCAC .......((.....((((.(((.(((........)))..))).))))(((((............))))))).......(((((........)))))........................ (-25.49 = -26.77 + 1.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:28 2006