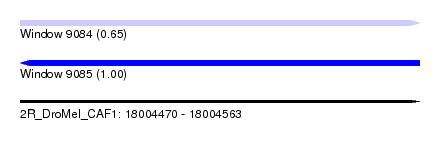
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,004,470 – 18,004,563 |
| Length | 93 |
| Max. P | 0.995016 |

| Location | 18,004,470 – 18,004,563 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 89.26 |
| Mean single sequence MFE | -43.53 |
| Consensus MFE | -37.59 |
| Energy contribution | -37.90 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
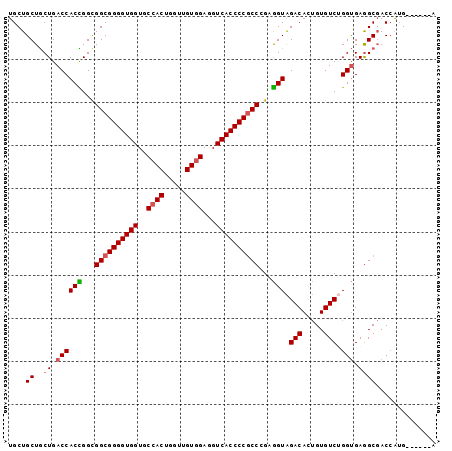
>2R_DroMel_CAF1 18004470 93 + 20766785 UGCUGCUGCUGACCACCGGUGGCGGGGUGGUGCCACUGGUUGUGGAAGUCACCCCGCCCGAGGUAGACACUGUGUCUGGUGAGGCGACCAUG------A ......((((.((((((...((((((((((..((((.....))))...))))))))))...)))((((.....)))))))..))))......------. ( -42.90) >DroSec_CAF1 18587 93 + 1 UGCUGCUGCUGACCACCGGCGGCGGGGUGGUGCCACUGGUUGUGGAGGUCACCCCGCCCGAGGUAGACACUGUGUCUGGUGAGGCGACCAUG------A ......((((.((((((...((((((((((..((((.....))))...))))))))))...)))((((.....)))))))..))))......------. ( -42.90) >DroSim_CAF1 17538 93 + 1 UGCUGCUGCUGACCACCGGCGGCGGGGUGGUGCCACUGGUUGUGGAGGUCACCCCGCCCGAGGUAGACACUGUGUCGGGUGAGGCGACCAUG------A ..(((((((((.....))))))))).......((((.....)))).((((.((.(((((((..(((...)))..))))))).)).))))...------. ( -45.20) >DroEre_CAF1 19907 93 + 1 UGCUGCUGCUGACCACCGGCGGCGGGGUGGUGCCACUGGUUGUAGAGGUCACCCCGCCGGAGGUAGACAUUGUGUCUGGUGAGGCGACCAUG------G ......((((.((((((..((((((((((..(((.(((....))).)))))))))))))..)))((((.....)))))))..))))......------. ( -42.50) >DroYak_CAF1 17215 93 + 1 UGCUGCUGCUGACCACUGGCGGCGGGGUGGUGCCACUGGUUGUGGAGGUCACCCCGCCGGAGGUAGACACUGUGUCUGGUGAGGCGACCAUG------G ......((((.((((((..(((((((((((..((((.....))))...)))))))))))..)))((((.....)))))))..))))......------. ( -44.30) >DroAna_CAF1 14406 99 + 1 UGCUGCUACUGACCACUGGCGGCGGGGUGGUGCCACUUGUGGUGGAUGUCACCCCACCGGAAGUGGACACAGUGUCGGGCGAAGCCACAGAGGAAAUAG ..(((.(((((.(((((..(((.((((((..(((((.....)))).)..)))))).)))..)))))...)))))...(((...))).)))......... ( -43.40) >consensus UGCUGCUGCUGACCACCGGCGGCGGGGUGGUGCCACUGGUUGUGGAGGUCACCCCGCCCGAGGUAGACACUGUGUCUGGUGAGGCGACCAUG______A ....((..((.((((((...((((((((((..((((.....))))...))))))))))...))).(((.....))).))).)))).............. (-37.59 = -37.90 + 0.31)

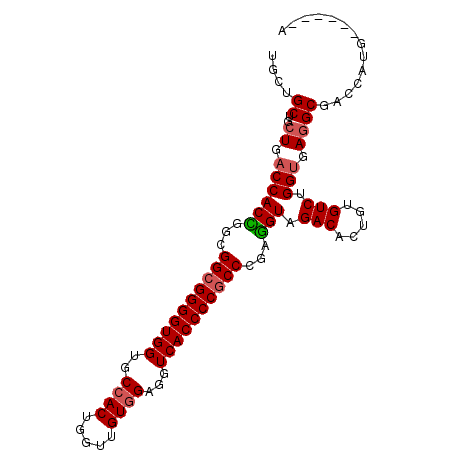
| Location | 18,004,470 – 18,004,563 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 89.26 |
| Mean single sequence MFE | -40.75 |
| Consensus MFE | -35.72 |
| Energy contribution | -35.75 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
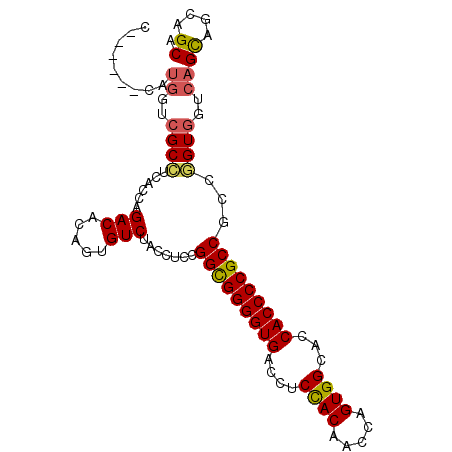
>2R_DroMel_CAF1 18004470 93 - 20766785 U------CAUGGUCGCCUCACCAGACACAGUGUCUACCUCGGGCGGGGUGACUUCCACAACCAGUGGCACCACCCCGCCACCGGUGGUCAGCAGCAGCA .------..((..((((.....((((.....))))......(((((((((....((((.....))))...)))))))))...))))..))((....)). ( -41.30) >DroSec_CAF1 18587 93 - 1 U------CAUGGUCGCCUCACCAGACACAGUGUCUACCUCGGGCGGGGUGACCUCCACAACCAGUGGCACCACCCCGCCGCCGGUGGUCAGCAGCAGCA .------..((..((((.....((((.....))))......(((((((((....((((.....))))...)))))))))...))))..))((....)). ( -41.30) >DroSim_CAF1 17538 93 - 1 U------CAUGGUCGCCUCACCCGACACAGUGUCUACCUCGGGCGGGGUGACCUCCACAACCAGUGGCACCACCCCGCCGCCGGUGGUCAGCAGCAGCA .------..((..((((......(((.....))).......(((((((((....((((.....))))...)))))))))...))))..))((....)). ( -40.20) >DroEre_CAF1 19907 93 - 1 C------CAUGGUCGCCUCACCAGACACAAUGUCUACCUCCGGCGGGGUGACCUCUACAACCAGUGGCACCACCCCGCCGCCGGUGGUCAGCAGCAGCA .------..((..((((.....((((.....)))).....((((((((((....((((.....))))...))))))))))..))))..))((....)). ( -40.40) >DroYak_CAF1 17215 93 - 1 C------CAUGGUCGCCUCACCAGACACAGUGUCUACCUCCGGCGGGGUGACCUCCACAACCAGUGGCACCACCCCGCCGCCAGUGGUCAGCAGCAGCA .------..((..(((......((((.....)))).....((((((((((....((((.....))))...))))))))))...)))..))((....)). ( -38.90) >DroAna_CAF1 14406 99 - 1 CUAUUUCCUCUGUGGCUUCGCCCGACACUGUGUCCACUUCCGGUGGGGUGACAUCCACCACAAGUGGCACCACCCCGCCGCCAGUGGUCAGUAGCAGCA .........(((((((...)))....((((...(((((..((((((((((....((((.....))))...))))))))))..))))).)))).)))).. ( -42.40) >consensus C______CAUGGUCGCCUCACCAGACACAGUGUCUACCUCCGGCGGGGUGACCUCCACAACCAGUGGCACCACCCCGCCGCCGGUGGUCAGCAGCAGCA .........((..((((......(((.....))).......(((((((((....((((.....))))...)))))))))...))))..))((....)). (-35.72 = -35.75 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:27 2006