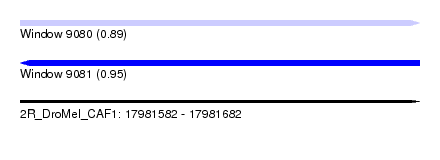
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,981,582 – 17,981,682 |
| Length | 100 |
| Max. P | 0.952526 |

| Location | 17,981,582 – 17,981,682 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -16.86 |
| Energy contribution | -17.78 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
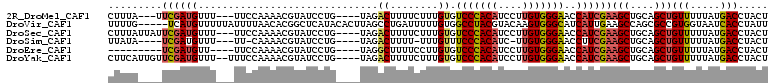
>2R_DroMel_CAF1 17981582 100 + 20766785 AGUAGGUCAUAAAAACAGCUGCAGCUUCGAUGGUUCCCACAAGGAUGUGGGACACAAAGAAAAGUCUA----CAGGAUACGUUUUGGAA---AAACAUCGAA---UAAAG .((((.(.........).))))...(((((((..(((((((....)))))))..(((((....(((..----...)))...)))))...---...)))))))---..... ( -24.50) >DroVir_CAF1 3104 105 + 1 AAUAGGUGAUUACCACGGCGCUGGCUUCAAUGAUGCCCACUUGUACGUAGGCCACAAAAAUCAGGCUAAGUGUAUGAGCCGUGUUAAAAUAAAAACAUGA-----CAAAA .........(((.((((((...(((.........)))...(..((((..((((..........))))...))))..))))))).))).............-----..... ( -23.40) >DroSec_CAF1 2954 103 + 1 AGUAGGUCAUAAAAACAGCUGCAGCUUCGAUGGUUCCCACAAGGAUGUGGGACACAAAGAAAAGUCUA----CAGGAUACGUUUUGGAA---AAACAUCGAAUAAUAAAG .((((.(.........).))))...(((((((..(((((((....)))))))..(((((....(((..----...)))...)))))...---...)))))))........ ( -24.50) >DroSim_CAF1 4118 96 + 1 AGUAGGUCAUAAAAACAGCUGCAGCUUCGAAGGUUCCCACAA-GAUGUGGGAAACAAA-AAAAGUCUA----CAGGAUACGUUUUG-AA---AAACAUCGA----UAUAA .((((.(.........).))))....((((.(.(((((((..-...))))))).((((-(...(((..----...)))...)))))-..---...).))))----..... ( -20.80) >DroEre_CAF1 2932 93 + 1 AGUAGGUCAUAAAAACAGCUGCAGCUUCGAUGGUUCCCACAAGGAUGUGGGACACAAGGAAAAGCCUA----CAGGAUACGUUUUGGAA----AACAUCGA--------- .((((.(.........).))))....((((((..(((((((....)))))))..(((((.....((..----..)).....)))))...----..))))))--------- ( -24.10) >DroYak_CAF1 3033 104 + 1 AGUAGGUCAUAAAAACAGCUGCAGCUUCGAUGGUUCCCACAAGGAUGUGGGACACAAAGAAAAGUCUA----CAGGAUACGUUUUGGAAA--AAACAUCGAACAAUGAAG ......((((......(((....)))((((((..(((((((....)))))))..(((((....(((..----...)))...)))))....--...))))))...)))).. ( -25.20) >consensus AGUAGGUCAUAAAAACAGCUGCAGCUUCGAUGGUUCCCACAAGGAUGUGGGACACAAAGAAAAGUCUA____CAGGAUACGUUUUGGAA___AAACAUCGA____UAAAG ................(((....)))((((((..((((((......))))))..(((((.....(((......))).....))))).........))))))......... (-16.86 = -17.78 + 0.92)

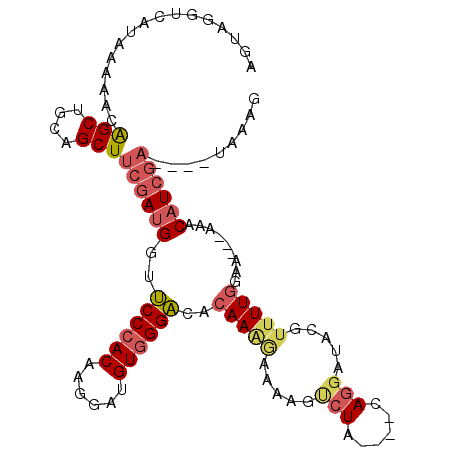
| Location | 17,981,582 – 17,981,682 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -15.79 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17981582 100 - 20766785 CUUUA---UUCGAUGUUU---UUCCAAAACGUAUCCUG----UAGACUUUUCUUUGUGUCCCACAUCCUUGUGGGAACCAUCGAAGCUGCAGCUGUUUUUAUGACCUACU .....---....((((((---.....))))))...(((----(((.((((....((..(((((((....)))))))..))..)))))))))).................. ( -25.10) >DroVir_CAF1 3104 105 - 1 UUUUG-----UCAUGUUUUUAUUUUAACACGGCUCAUACACUUAGCCUGAUUUUUGUGGCCUACGUACAAGUGGGCAUCAUUGAAGCCAGCGCCGUGGUAAUCACCUAUU ....(-----((.((((........)))).))).....(((...(((((..(((.(((((((((......))))))..))).)))..))).)).)))............. ( -24.60) >DroSec_CAF1 2954 103 - 1 CUUUAUUAUUCGAUGUUU---UUCCAAAACGUAUCCUG----UAGACUUUUCUUUGUGUCCCACAUCCUUGUGGGAACCAUCGAAGCUGCAGCUGUUUUUAUGACCUACU ............((((((---.....))))))...(((----(((.((((....((..(((((((....)))))))..))..)))))))))).................. ( -25.10) >DroSim_CAF1 4118 96 - 1 UUAUA----UCGAUGUUU---UU-CAAAACGUAUCCUG----UAGACUUUU-UUUGUUUCCCACAUC-UUGUGGGAACCUUCGAAGCUGCAGCUGUUUUUAUGACCUACU (((((----...((((((---..-..))))))...(((----(((.((((.-...(.(((((((...-..))))))).)...)))))))))).......)))))...... ( -23.50) >DroEre_CAF1 2932 93 - 1 ---------UCGAUGUU----UUCCAAAACGUAUCCUG----UAGGCUUUUCCUUGUGUCCCACAUCCUUGUGGGAACCAUCGAAGCUGCAGCUGUUUUUAUGACCUACU ---------.....(((----....((((((....(((----(((.((((....((..(((((((....)))))))..))..)))))))))).))))))...)))..... ( -25.50) >DroYak_CAF1 3033 104 - 1 CUUCAUUGUUCGAUGUUU--UUUCCAAAACGUAUCCUG----UAGACUUUUCUUUGUGUCCCACAUCCUUGUGGGAACCAUCGAAGCUGCAGCUGUUUUUAUGACCUACU ..((((......((((((--(....)))))))...(((----(((.((((....((..(((((((....)))))))..))..))))))))))........))))...... ( -26.80) >consensus CUUUA____UCGAUGUUU___UUCCAAAACGUAUCCUG____UAGACUUUUCUUUGUGUCCCACAUCCUUGUGGGAACCAUCGAAGCUGCAGCUGUUUUUAUGACCUACU .........((((((..............................((........)).(((((((....)))))))..))))))(((....)))(((.....)))..... (-15.79 = -15.85 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:24 2006