| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,971,622 – 17,971,722 |
| Length | 100 |
| Max. P | 0.997896 |

| Location | 17,971,622 – 17,971,722 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 71.57 |
| Mean single sequence MFE | -29.21 |
| Consensus MFE | -16.41 |
| Energy contribution | -18.50 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
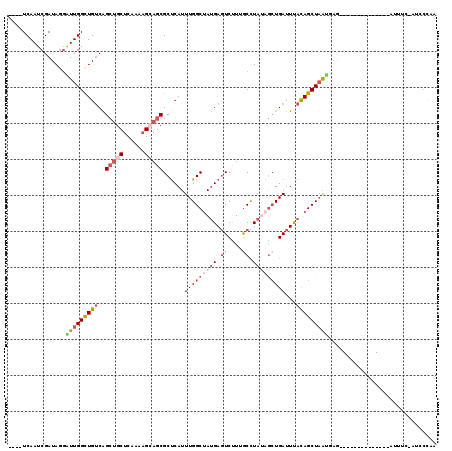
>2R_DroMel_CAF1 17971622 100 + 20766785 ----UCAAUCGAUAGGAUUGGCUGACGGCUGCUCAAAAGCAGCGCUCAUUUGGCUACGAGUCUUUACCUAUAGCUGAUUUACAGCUAAUGCG--------------AUUCC-AUGCUAA ----..(((((.(((((..((((..((((((((....))))))(((.....)))..))))))..).))))((((((.....))))))...))--------------)))..-....... ( -29.30) >DroSec_CAF1 28781 100 + 1 ----UCAAUCGAUAGGGUUGGCUGUCAGCUGCUCAAAAGCAGCGCUCAUUUGGCUAUGAGUCCUUGCCUAUAGCUGAUUUACAGCUAAUGAG--------------AUUUC-AUCCCAA ----..........(((.(((......((((((....)))))).((((((.((((.((((((...((.....)).)))))).))))))))))--------------...))-).))).. ( -32.80) >DroSim_CAF1 24403 100 + 1 ----UCAAUCGAUAGGGUUGGCUGUCAGCUGCUCAAAAGCAGCGCUCAUUUGGCUAUGAGUCUUUGCCUAUAGCUGAUUUACAGCUAAUGAG--------------AUUUU-AUCCCAA ----......((((((((((((.....((((((....))))))((((((......))))))....)))..((((((.....))))))....)--------------)))))-))).... ( -32.80) >DroEre_CAF1 33195 102 + 1 ----UCAAUCGAUACGAUUGGCUGUCAGCUG-----------CGCCCAUUUGGCUAUGAUUCUUAGCUUA--GCUGAUUCAUAGCUUGAAAACCGCACAAAUGGUCAUAUGUUUCCCAA ----......((.(((..(((((((..(.((-----------((.......((((((((...(((((...--))))).)))))))).......)))))..)))))))..))).)).... ( -27.24) >DroYak_CAF1 33307 97 + 1 AUACUCAAUCGAUAGCUUUGGUUGUCAGCUGCACAAAAGAAGCGCUCAUUUGGCCAUGAGUCUUGGCCUA--AAUGAUUUACAGCUAAAUAA------AA------AUAUG-------- ..........((((((....))))))((((((.(.......).))(((((((((((.......))))).)--))))).....))))......------..------.....-------- ( -23.90) >consensus ____UCAAUCGAUAGGAUUGGCUGUCAGCUGCUCAAAAGCAGCGCUCAUUUGGCUAUGAGUCUUUGCCUAUAGCUGAUUUACAGCUAAUGAG______________AUUUC_AUCCCAA ................(((((((((..(((((......)))))......(((((((((.((....)).)))))))))...))))))))).............................. (-16.41 = -18.50 + 2.09)

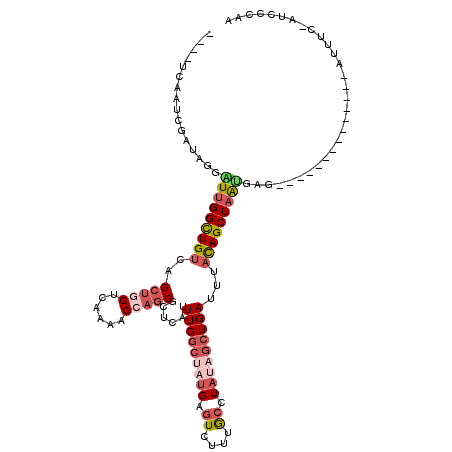
| Location | 17,971,622 – 17,971,722 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 71.57 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -12.91 |
| Energy contribution | -14.96 |
| Covariance contribution | 2.05 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
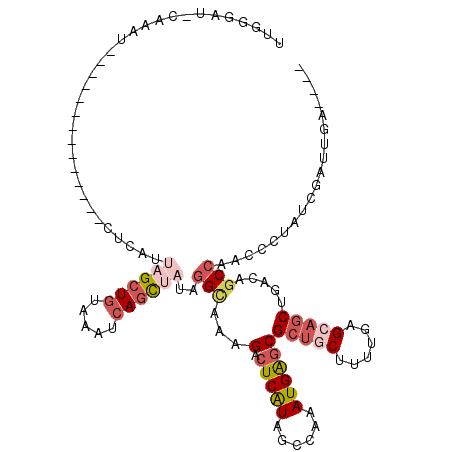
>2R_DroMel_CAF1 17971622 100 - 20766785 UUAGCAU-GGAAU--------------CGCAUUAGCUGUAAAUCAGCUAUAGGUAAAGACUCGUAGCCAAAUGAGCGCUGCUUUUGAGCAGCCGUCAGCCAAUCCUAUCGAUUGA---- .......-..(((--------------((...((((((.....))))))..(((...(.(((((......))))))((((((....)))))).....)))........)))))..---- ( -27.10) >DroSec_CAF1 28781 100 - 1 UUGGGAU-GAAAU--------------CUCAUUAGCUGUAAAUCAGCUAUAGGCAAGGACUCAUAGCCAAAUGAGCGCUGCUUUUGAGCAGCUGACAGCCAACCCUAUCGAUUGA---- ..(((((-((...--------------.))))((((((.....))))))..(((...(.(((((......))))))((((((....)))))).....)))..)))..........---- ( -31.60) >DroSim_CAF1 24403 100 - 1 UUGGGAU-AAAAU--------------CUCAUUAGCUGUAAAUCAGCUAUAGGCAAAGACUCAUAGCCAAAUGAGCGCUGCUUUUGAGCAGCUGACAGCCAACCCUAUCGAUUGA---- ..(((..-.....--------------.....((((((.....))))))..(((...(.(((((......))))))((((((....)))))).....)))..)))..........---- ( -31.70) >DroEre_CAF1 33195 102 - 1 UUGGGAAACAUAUGACCAUUUGUGCGGUUUUCAAGCUAUGAAUCAGC--UAAGCUAAGAAUCAUAGCCAAAUGGGCG-----------CAGCUGACAGCCAAUCGUAUCGAUUGA---- ...(....).......((.((((((((((.....(((((((.(((((--...)))..)).)))))))......(((.-----------.........)))))))))).))).)).---- ( -24.60) >DroYak_CAF1 33307 97 - 1 --------CAUAU------UU------UUAUUUAGCUGUAAAUCAUU--UAGGCCAAGACUCAUGGCCAAAUGAGCGCUUCUUUUGUGCAGCUGACAACCAAAGCUAUCGAUUGAGUAU --------.((((------((------.....(((((.....(((((--(.(((((.......)))))))))))((((.......)))).............)))))......)))))) ( -25.30) >consensus UUGGGAU_CAAAU______________CUCAUUAGCUGUAAAUCAGCUAUAGGCAAAGACUCAUAGCCAAAUGAGCGCUGCUUUUGAGCAGCUGACAGCCAACCCUAUCGAUUGA____ ................................((((((.....))))))..(((...(.(((((......))))))(((((......))))).....)))................... (-12.91 = -14.96 + 2.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:20 2006